| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,452,878 – 12,452,969 |
| Length | 91 |
| Max. P | 0.991698 |

| Location | 12,452,878 – 12,452,969 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 69.74 |
| Shannon entropy | 0.62521 |
| G+C content | 0.49783 |
| Mean single sequence MFE | -23.39 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
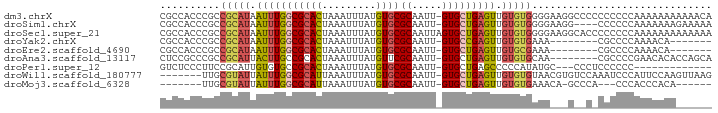
>dm3.chrX 12452878 91 - 22422827 CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGGGGAAGGCCCCCCCCCCAAAAAAAAAAACA .(((..((.((((((((((..((((((.........))))((....-))))..))))))))))))..)))...................... ( -32.60, z-score = -2.81, R) >droSim1.chrX 9571779 87 - 17042790 CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGGGGAAGG----CCCCCCAAAAAAAGAAAAA .(((..((.((((((((((..((((((.........))))((....-))))..))))))))))))..))----).................. ( -32.60, z-score = -3.06, R) >droSec1.super_21 208295 92 - 1102487 CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUUAGUGCUGAGUUGUGUGGGGAAGGCACCCCCCCCAAAAAAAAAAAAA .(((..((.((((((((((..((((((.........))))((.....))))..))))))))))))..)))...................... ( -33.20, z-score = -3.15, R) >droYak2.chrX 6728887 76 - 21770863 CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCCGAGUUGUGUGAAA--------CGCCCCAAAACA------- ..........(((((((((((((((((.........))))((....-)))))))))))))))...--------............------- ( -23.70, z-score = -2.03, R) >droEre2.scaffold_4690 4341907 76 + 18748788 CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGCGAAA--------CGCCCCAAAACA------- ..........(((((((((..((((((.........))))((....-))))..)))))))))...--------............------- ( -23.10, z-score = -1.60, R) >droAna3.scaffold_13117 3338857 83 - 5790199 CUCCGCCCGCCGCAUUACUUGCCGCACUAAAUUUAUGUUCGCAAUU-GUGCUGAGUUGUGUGCAA--------CGCCCCGAACACACCAGCA ....((..((.(((.....))).))..........((((((((...-.)))...((((....)))--------).....))))).....)). ( -18.50, z-score = -0.26, R) >droPer1.super_12 2355062 75 + 2414086 GUCUCCCUUCCGCAUUGUGUGCCGCACUAAAUUUAUGUGCGCAAUU-GUGCUGAGCCCCCAUAUGC---CCCUCCCCCC------------- .......(((.((((....(((.((((.........)))))))...-)))).)))...........---..........------------- ( -14.70, z-score = -2.70, R) >droWil1.scaffold_180777 4253114 84 - 4753960 -------UUGCGUAUUAUUUGGCGCAUUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGUAACGUGUCCAAAUCCCAUUCCAAGUUAAG -------.(((((........))))).........((..(((((((-......)))))))..))............................ ( -15.00, z-score = 0.43, R) >droMoj3.scaffold_6328 1994394 74 - 4453435 -------UUGCGUAUUAUUUGGCGCAUUAAAUUUAUGUGCGCAAUU-GUGCUGAGUUGUGUGAAACA-GCCCA---CCCACCCACA------ -------....((((...(((((((((.......)))))).)))..-))))((.(((((.....)))-)).))---..........------ ( -17.10, z-score = -0.11, R) >consensus CGCCACCCGCCGCAUAAUUUGGCGCACUAAAUUUAUGUGCGCAAUU_GUGCUGAGUUGUGUGAAA___G__C_CCCCCCAAAAAAA______ ...........((((...(((.(((((.........))))))))...))))......................................... (-13.00 = -12.88 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:41 2011