| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,435,619 – 12,435,706 |
| Length | 87 |
| Max. P | 0.996132 |

| Location | 12,435,619 – 12,435,706 |
|---|---|
| Length | 87 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.50790 |
| G+C content | 0.43151 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.52 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
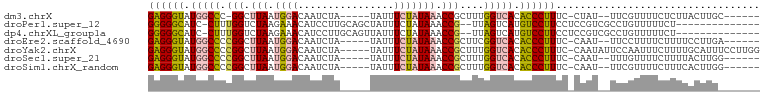
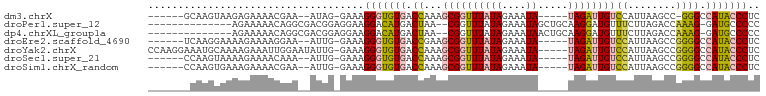
>dm3.chrX 12435619 87 + 22422827 GAGGGUAUGGCCC-GGCUUAAUGGACAAUCUA-----UAUUUCUAUAAACCGCUUUGGUCACACCCUUUC-CUAU--UUCGUUUUCUCUUACUUGC------ ((((((.((((((-((.((.(((((.......-----....))))).)))))....))))).))))))..-....--...................------ ( -19.40, z-score = -1.85, R) >droPer1.super_12 2323890 85 - 2414086 GGGGGCAUC-CUUUGGUCUAAGAAACAUCCUUGCAGCUAUUUCUAUAAACCG--UUAGUCAUGUCCUUCCUCCGUCGCCUGUUUUUCU-------------- ((((((((.-((.((((.(((((((..............))))).)).))))--..))..))))))))....................-------------- ( -12.54, z-score = 0.93, R) >dp4.chrXL_group1a 3218918 85 + 9151740 GGGGGCAUC-CUUUGGUCUAAGAAACAUCCUUGCAGUUAUUUCUAUAAACCG--UUAGUCAUGUCCUUCCUCCGUCGCCUGUUUUUCU-------------- ((((((((.-((.((((.(((((((...(......)...))))).)).))))--..))..))))))))....................-------------- ( -12.90, z-score = 0.55, R) >droEre2.scaffold_4690 4324845 88 - 18748788 GAGGGUAUGGCCCCGGCUUAAUGGACAAUCUA-----UAUUUCUAUAAACCGCUUCGGUCACACCCUUUC-CAAU--UUCCUUUUCUUUUCCUUGA------ ((((((.(((((.(((.((.(((((.......-----....))))).)))))....))))).))))))..-....--...................------ ( -19.70, z-score = -1.69, R) >droYak2.chrX 6711915 96 + 21770863 GAGGGUAUGGCCCCGGCUUAAUGGACAAUCUA-----UAUUUCUAUAAACCGCUUUGGUCACACCCUUUC-CAAUAUUCCAAUUUCUUUUGCAUUUCCUUGG ((((((.(((((.(((.((.(((((.......-----....))))).)))))....))))).)))))).(-(((.....(((......))).......)))) ( -20.40, z-score = -1.09, R) >droSec1.super_21 191518 88 + 1102487 GAGGGUAUGGCCCCGGCUUAAUGGACAAUCUA-----UAUUUCUAUAAACCGCUUUGGUCACACCCUUUC-CAAU--UUUGUUUUCUUUUACUUGG------ ((((((.(((((.(((.((.(((((.......-----....))))).)))))....))))).))))))..-....--...................------ ( -19.70, z-score = -1.44, R) >droSim1.chrX_random 3379882 88 + 5698898 GAGGGUAUGGCCCCGGCUUAAUGGACAAUCUA-----UAUUUCUAUAAACCGCUUUGGUCACACCCUUUC-CAAU--UUCGUUUUCUUUCACUUGG------ ((((((.(((((.(((.((.(((((.......-----....))))).)))))....))))).))))))..-....--...................------ ( -19.70, z-score = -1.50, R) >consensus GAGGGUAUGGCCCCGGCUUAAUGGACAAUCUA_____UAUUUCUAUAAACCGCUUUGGUCACACCCUUUC_CAAU__UUCGUUUUCUUUUACUUG_______ ((((((.(((((.(((.(((.((((................))))))).)))....))))).)))))).................................. (-13.53 = -13.52 + -0.01)
| Location | 12,435,619 – 12,435,706 |
|---|---|
| Length | 87 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.50790 |
| G+C content | 0.43151 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -14.82 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
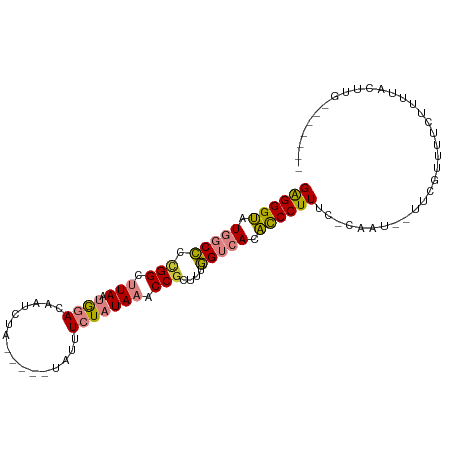
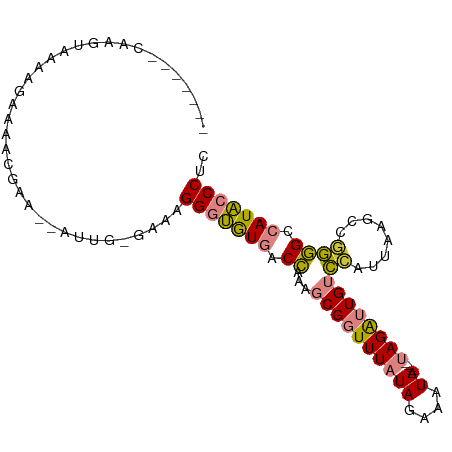
>dm3.chrX 12435619 87 - 22422827 ------GCAAGUAAGAGAAAACGAA--AUAG-GAAAGGGUGUGACCAAAGCGGUUUAUAGAAAUA-----UAGAUUGUCCAUUAAGCC-GGGCCAUACCCUC ------...................--....-...((((((((.((...((((((((((....))-----))))))))..........-.)).)))))))). ( -21.64, z-score = -2.37, R) >droPer1.super_12 2323890 85 + 2414086 --------------AGAAAAACAGGCGACGGAGGAAGGACAUGACUAA--CGGUUUAUAGAAAUAGCUGCAAGGAUGUUUCUUAGACCAAAG-GAUGCCCCC --------------.........((((.(...(..((.......))..--)((((((.(((((((.((....)).)))))))))))))....-).))))... ( -18.20, z-score = -0.83, R) >dp4.chrXL_group1a 3218918 85 - 9151740 --------------AGAAAAACAGGCGACGGAGGAAGGACAUGACUAA--CGGUUUAUAGAAAUAACUGCAAGGAUGUUUCUUAGACCAAAG-GAUGCCCCC --------------.........((((.(...(..((.......))..--)((((((.(((((((.((....)).)))))))))))))....-).))))... ( -18.20, z-score = -1.14, R) >droEre2.scaffold_4690 4324845 88 + 18748788 ------UCAAGGAAAAGAAAAGGAA--AUUG-GAAAGGGUGUGACCGAAGCGGUUUAUAGAAAUA-----UAGAUUGUCCAUUAAGCCGGGGCCAUACCCUC ------...................--....-...((((((((.((...((((((((((....))-----))))))))((........)))).)))))))). ( -24.70, z-score = -2.44, R) >droYak2.chrX 6711915 96 - 21770863 CCAAGGAAAUGCAAAAGAAAUUGGAAUAUUG-GAAAGGGUGUGACCAAAGCGGUUUAUAGAAAUA-----UAGAUUGUCCAUUAAGCCGGGGCCAUACCCUC ((((.....(.(((......))).)...)))-)..((((((((.((...((((((((((....))-----))))))))((........)))).)))))))). ( -27.40, z-score = -2.45, R) >droSec1.super_21 191518 88 - 1102487 ------CCAAGUAAAAGAAAACAAA--AUUG-GAAAGGGUGUGACCAAAGCGGUUUAUAGAAAUA-----UAGAUUGUCCAUUAAGCCGGGGCCAUACCCUC ------((((...............--.)))-)..((((((((.((...((((((((((....))-----))))))))((........)))).)))))))). ( -26.19, z-score = -3.47, R) >droSim1.chrX_random 3379882 88 - 5698898 ------CCAAGUGAAAGAAAACGAA--AUUG-GAAAGGGUGUGACCAAAGCGGUUUAUAGAAAUA-----UAGAUUGUCCAUUAAGCCGGGGCCAUACCCUC ------((((.((........))..--.)))-)..((((((((.((...((((((((((....))-----))))))))((........)))).)))))))). ( -27.10, z-score = -3.53, R) >consensus _______CAAGUAAAAGAAAACGAA__AUUG_GAAAGGGUGUGACCAAAGCGGUUUAUAGAAAUA_____UAGAUUGUCCAUUAAGCCGGGGCCAUACCCUC ....................................(((((((.((.....((((((.........................))))))..)).))))))).. (-14.82 = -13.97 + -0.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:38 2011