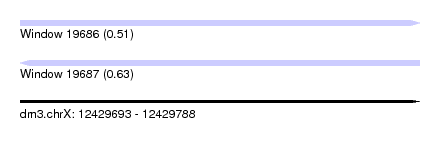
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,429,693 – 12,429,788 |
| Length | 95 |
| Max. P | 0.625894 |

| Location | 12,429,693 – 12,429,788 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.37 |
| Shannon entropy | 0.28968 |
| G+C content | 0.53536 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -22.60 |
| Energy contribution | -24.20 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12429693 95 + 22422827 -GGACUACGGAUUGUGGGCUAGGUCAGCCAACA---UCCGAGGCCAGUGGCCCACUGGACAUUAUCUAAUCCGCGUCCGUCCG-CUUUUUGCCCGUUUUU -((((..((((((((((((((((((........---.....))))..)))))))).(((.....))))))))).))))....(-(.....))........ ( -30.62, z-score = -0.88, R) >droSim1.chrX 9559926 99 + 17042790 UGGACUGCGGAUUGUGGGCUAGGUCAGCCAACAACAUCCGAGGCCAGUG-CCCACUGGCCAUUAUCUAAUCCGCGCCAUUCCUUCUUUUUGCCGGCUUUU ......(((((((((.((((.....)))).)).........(((((((.-...))))))).......)))))))(((................))).... ( -33.89, z-score = -1.38, R) >droSec1.super_21 185854 98 + 1102487 UGGACUGCGGAUUGUGGGCUAGGUCAGCCAACAACAUCCGAGGCCAGUG-CCCACUGGCCAUUAUCUAAUCCGCGCCAUCCCUUCUUUUUGACGGUUUU- (((...(((((((((.((((.....)))).)).........(((((((.-...))))))).......))))))).)))..((.((.....)).))....- ( -35.30, z-score = -2.23, R) >droYak2.chrX 6706153 93 + 21770863 CGGACUGCAGAUUGUGGGCUAGGUCAGCCAACA---UCCGUGGCCAUUG-CCCACUGGCCAUUAUCUAAUCCGCAACCUGCU---UUUUUGCCGGUUUUU (((...((((.((((((..(((((.........---...(((((((...-.....))))))).)))))..)))))).)))).---......)))...... ( -31.22, z-score = -1.58, R) >droEre2.scaffold_4690 4318995 87 - 18748788 UGGACAGCGGAUUGUGGGCUAGGUCAGCCAACA---------GCCAUUG-CCCACUGAGCAUUAUCUAAUCCGCAACCUCCU---UUUUAGCCGGUUUUU ......(((((((((((((..(((.........---------)))...)-))))).((......)).)))))))((((..((---....))..))))... ( -26.40, z-score = -1.35, R) >consensus UGGACUGCGGAUUGUGGGCUAGGUCAGCCAACA___UCCGAGGCCAGUG_CCCACUGGCCAUUAUCUAAUCCGCGACCUCCC__CUUUUUGCCGGUUUUU ......(((((((((.((((.....)))).)).........((((((((...)))))))).......))))))).......................... (-22.60 = -24.20 + 1.60)

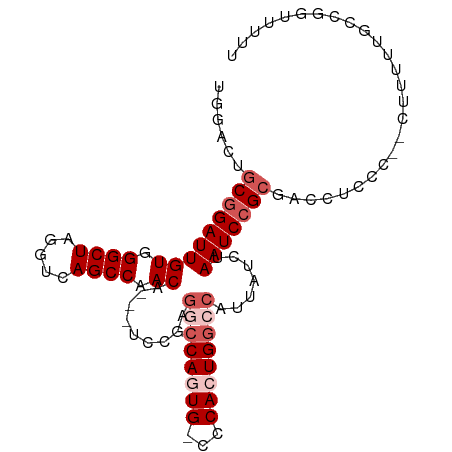
| Location | 12,429,693 – 12,429,788 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.37 |
| Shannon entropy | 0.28968 |
| G+C content | 0.53536 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -26.30 |
| Energy contribution | -27.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
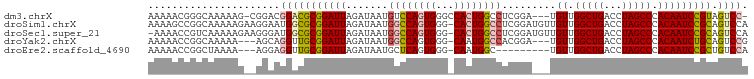
>dm3.chrX 12429693 95 - 22422827 AAAAACGGGCAAAAAG-CGGACGGACGCGGAUUAGAUAAUGUCCAGUGGGCCACUGGCCUCGGA---UGUUGGCUGACCUAGCCCACAAUCCGUAGUCC- ......((((.....(-((......)))((.((((.((((((((...(((((...))))).)))---))))).))))))..))))..............- ( -33.60, z-score = -1.49, R) >droSim1.chrX 9559926 99 - 17042790 AAAAGCCGGCAAAAAGAAGGAAUGGCGCGGAUUAGAUAAUGGCCAGUGGG-CACUGGCCUCGGAUGUUGUUGGCUGACCUAGCCCACAAUCCGCAGUCCA ....((((..............))))..(((((.......(((((((...-.))))))).(((((..(((.(((((...))))).)))))))).))))). ( -35.54, z-score = -1.10, R) >droSec1.super_21 185854 98 - 1102487 -AAAACCGUCAAAAAGAAGGGAUGGCGCGGAUUAGAUAAUGGCCAGUGGG-CACUGGCCUCGGAUGUUGUUGGCUGACCUAGCCCACAAUCCGCAGUCCA -....(((((..........)))))...(((((.......(((((((...-.))))))).(((((..(((.(((((...))))).)))))))).))))). ( -35.80, z-score = -1.57, R) >droYak2.chrX 6706153 93 - 21770863 AAAAACCGGCAAAAA---AGCAGGUUGCGGAUUAGAUAAUGGCCAGUGGG-CAAUGGCCACGGA---UGUUGGCUGACCUAGCCCACAAUCUGCAGUCCG ...((((.((.....---.)).)))).((((((......((((((.....-...))))))((((---(((.(((((...))))).)).))))).)))))) ( -33.10, z-score = -1.73, R) >droEre2.scaffold_4690 4318995 87 + 18748788 AAAAACCGGCUAAAA---AGGAGGUUGCGGAUUAGAUAAUGCUCAGUGGG-CAAUGGC---------UGUUGGCUGACCUAGCCCACAAUCCGCUGUCCA ...((((..((....---))..))))(((((((.((......)).(((((-(...(((---------........).))..)))))))))))))...... ( -27.60, z-score = -0.83, R) >consensus AAAAACCGGCAAAAAG__GGAAGGGCGCGGAUUAGAUAAUGGCCAGUGGG_CACUGGCCUCGGA___UGUUGGCUGACCUAGCCCACAAUCCGCAGUCCA ......................(((((((((((.......((((((((...)))))))).........((.(((((...))))).))))))))).)))). (-26.30 = -27.30 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:36 2011