| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,429,357 – 12,429,453 |
| Length | 96 |
| Max. P | 0.933137 |

| Location | 12,429,357 – 12,429,453 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Shannon entropy | 0.45691 |
| G+C content | 0.40662 |
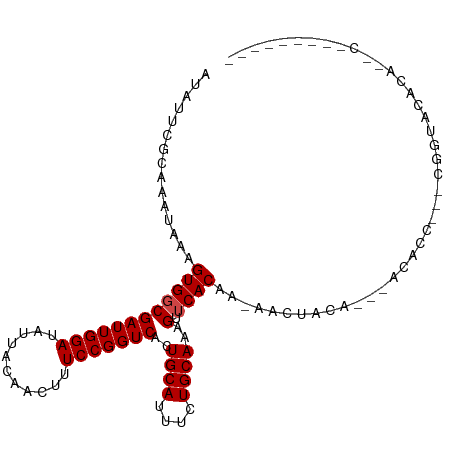
| Mean single sequence MFE | -16.69 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
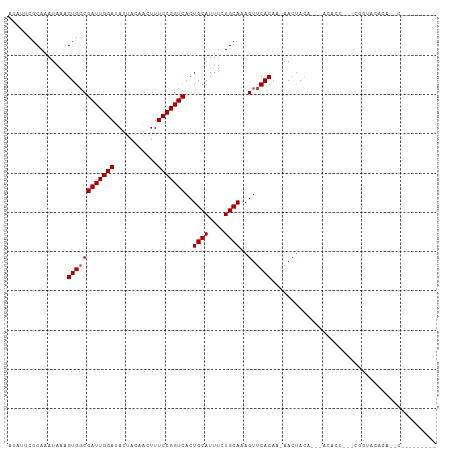
>dm3.chrX 12429357 96 - 22422827 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---GCACUCGACGGUACACAAGC--------- ....((((.......(..(.(((((((...........))))))).)..).....(((..((((.....-))))...---)))...).))).........--------- ( -16.20, z-score = -0.15, R) >droPer1.super_12 2316856 108 + 2414086 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACACCCCCCCCCCCCCCUCCACUCACCAUAAAAGC ..............(((((.(((((((...........)))))))..((((....))))..........-......................)))))............ ( -16.10, z-score = -1.18, R) >dp4.chrXL_group1a 3211882 103 - 9151740 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA-----CCCCCCCCCUCCACUUGCCAUAAAAGC ...............((((((((((((...........)))))))..((((....))))..........-.......-----................)))))...... ( -18.40, z-score = -1.49, R) >droAna3.scaffold_13117 3310950 77 - 5790199 AUGUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACAA------------------------------- .(((..(((...((.(..(.(((((((...........))))))).)..).))..)))..((((.....-))))))).------------------------------- ( -15.40, z-score = -1.39, R) >droEre2.scaffold_4690 4318666 86 + 18748788 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAGCUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---ACACU---CGGUA---------------- ......(((...((.(..(.(((((((...........))))))).)..).))..)))..((((.....-))))...---.....---.....---------------- ( -15.30, z-score = -0.65, R) >droYak2.chrX 6705781 93 - 21770863 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---ACACU---CGGUGUACAAGC--------- ......(((...((.(..(.(((((((...........))))))).)..).))..)))...((.(((..-.......---.....---..))).))....--------- ( -16.69, z-score = -0.66, R) >droSec1.super_21 185513 93 - 1102487 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---ACACU---CGGUACACAAGC--------- ......(((...((.(..(.(((((((...........))))))).)..).))..)))..((((.....-))))...---.....---............--------- ( -15.30, z-score = -0.78, R) >droSim1.chrX 9559599 93 - 17042790 AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---ACACU---CGGUACACAAGC--------- ......(((...((.(..(.(((((((...........))))))).)..).))..)))..((((.....-))))...---.....---............--------- ( -15.30, z-score = -0.78, R) >droWil1.scaffold_180777 4226624 76 - 4753960 CAACUCGCAAAUAAAGUGCCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AAACUAU-------------------------------- ......(((....((((((.(((((((...........)))))))...)))))).)))..((((.....-.))))..-------------------------------- ( -14.90, z-score = -2.25, R) >droMoj3.scaffold_6328 1956226 103 - 4453435 CAGUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AAAAAAA---AGAACUACAAAUGCAAACAACAUGAGG-- ..(((.(((......(..(.(((((((...........))))))).)..)..........(((((....-.......---.))))).....))).))).........-- ( -20.90, z-score = -1.91, R) >droGri2.scaffold_14853 2585149 95 + 10151454 GAGCUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCAAUGCAUUUCUGCAAAGUUCACAACAACUACA---ACAACUUACCACCCCCCC----------- ((((((((.......)))..(((((((...........)))))))..((((....)))).)))))............---..................----------- ( -15.90, z-score = -0.93, R) >droVir3.scaffold_12928 980343 84 - 7717345 GGGCUUGCAAAUAAAGUGCCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA-AACUACA---AAUACCAG--------------------- (((((((((....((((((.(((((((...........)))))))...)))))).))).))))))....-.......---........--------------------- ( -19.90, z-score = -2.49, R) >consensus AUAUUCGCAAAUAAAGUGGCGAUUGGAUAUUACAACUUUCCGGUCACUGCAUUUCUGCAAAGUUCACAA_AACUACA___ACACC___CGGUACACA__C_________ ...............((((((((((((...........)))))))..((((....))))..).)))).......................................... (-13.48 = -13.65 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:35 2011