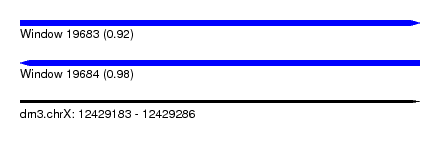
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,429,183 – 12,429,286 |
| Length | 103 |
| Max. P | 0.983864 |

| Location | 12,429,183 – 12,429,286 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.48380 |
| G+C content | 0.50804 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920359 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
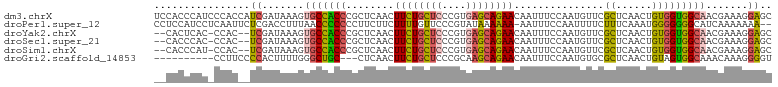
>dm3.chrX 12429183 103 + 22422827 GCUCCUUUCGUUGCCACCACAGUUGAGCGAACAUUGGAAAUUGUUCUGCUCACGGGAGCAGAAGUUGAGCGGGUGGCACUUUAUCGAUGGUGGGAUGGGUGGA ..(((.((((((((((((...(((.((((((((.(....).)))))(((((....)))))...))).))).))))))(((........)))..)))))).))) ( -37.10, z-score = -1.55, R) >droPer1.super_12 2316709 100 - 2414086 --UUUUUUUGAUGCCCCCCCAUUUGAAAGAAAAUUGGAAAUU-UUUUUUAUACGGGAACAAAAGAAGAAGGGGGGGUUAAAGGUCGAGAAUUGAGGAUGGAGG --..((((((((((((((((.....(((((((((.....)))-))))))....(....)..........)))))))).....))))))))............. ( -27.30, z-score = -1.76, R) >droYak2.chrX 6705575 98 + 21770863 GCUCCUUUCGUUGCCACCACAGUUGAGCGAACAUUGGAAAUUGUUCUGCUCACGGGAGCAGAAGUUGAGCGGGUGGCACUUUAUCGA--GUGG-GUGAGUG-- ((((((((((.(((((((...(((.((((((((.(....).)))))(((((....)))))...))).))).)))))))......)))--)..)-).)))).-- ( -36.90, z-score = -2.08, R) >droSec1.super_21 185346 98 + 1102487 GCUCCUUUCGUUGCCACCACAGUUGAGCGAACAUUGGAAAUUGUUCUGCUCACGGGAGCAGAAGUUGAGCGGGUGGCACUUUAUCGA--GUGG-GUGGGUG-- ((.((.((((.(((((((...(((.((((((((.(....).)))))(((((....)))))...))).))).)))))))......)))--).))-)).....-- ( -36.30, z-score = -1.72, R) >droSim1.chrX 9559445 98 + 17042790 GCUCCUUUCGUUGCCACCACAGUUGAGCGAACAUUGGAAAUUGUUCUGCUCACGGGAGCAGAAGUUGAGCGGGUGGCACUUUAUCGA--GUGG-AUGGGUG-- ..(((.((((.(((((((...(((.((((((((.(....).)))))(((((....)))))...))).))).)))))))......)))--).))-)......-- ( -36.70, z-score = -2.10, R) >droGri2.scaffold_14853 2585017 90 - 10151454 ACCCCUUUGUUUGCCACUACAGUUGAGCGCACAUUGGAAAUUGUUCUGCUUGCGGGAGCAGAAGUUGAG---GCAGCCCAAAAGUGGGGAAGG---------- ...(((((..(((((....(((((...(........).)))))((((((((....)))))))).....)---))))((((....)))))))))---------- ( -26.10, z-score = 0.44, R) >consensus GCUCCUUUCGUUGCCACCACAGUUGAGCGAACAUUGGAAAUUGUUCUGCUCACGGGAGCAGAAGUUGAGCGGGUGGCACUUUAUCGA__GUGG_GUGGGUG__ ............((((((...(((.(((..(((........)))(((((((....))))))).))).))).)))))).......................... (-21.43 = -21.60 + 0.17)
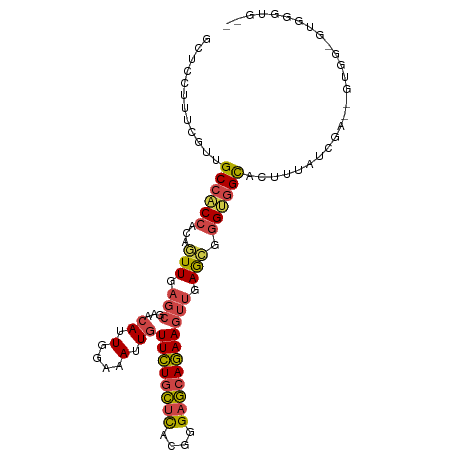
| Location | 12,429,183 – 12,429,286 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.48380 |
| G+C content | 0.50804 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -17.71 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983864 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
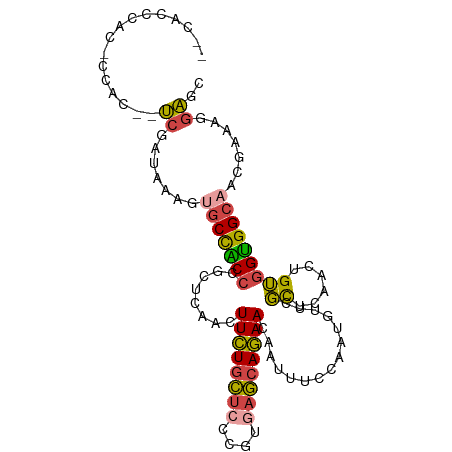
>dm3.chrX 12429183 103 - 22422827 UCCACCCAUCCCACCAUCGAUAAAGUGCCACCCGCUCAACUUCUGCUCCCGUGAGCAGAACAAUUUCCAAUGUUCGCUCAACUGUGGUGGCAACGAAAGGAGC (((....(((........)))....((((((((((.........)).....(((((.(((((........))))))))))...).)))))))......))).. ( -28.00, z-score = -2.28, R) >droPer1.super_12 2316709 100 + 2414086 CCUCCAUCCUCAAUUCUCGACCUUUAACCCCCCCUUCUUCUUUUGUUCCCGUAUAAAAAA-AAUUUCCAAUUUUCUUUCAAAUGGGGGGGCAUCAAAAAAA-- ...........................(((((((......((((((......))))))((-(((.....))))).........)))))))...........-- ( -16.60, z-score = -1.00, R) >droYak2.chrX 6705575 98 - 21770863 --CACUCAC-CCAC--UCGAUAAAGUGCCACCCGCUCAACUUCUGCUCCCGUGAGCAGAACAAUUUCCAAUGUUCGCUCAACUGUGGUGGCAACGAAAGGAGC --.......-...(--((.......((((((((((.........)).....(((((.(((((........))))))))))...).))))))).(....)))). ( -29.30, z-score = -2.37, R) >droSec1.super_21 185346 98 - 1102487 --CACCCAC-CCAC--UCGAUAAAGUGCCACCCGCUCAACUUCUGCUCCCGUGAGCAGAACAAUUUCCAAUGUUCGCUCAACUGUGGUGGCAACGAAAGGAGC --.......-...(--((.......((((((((((.........)).....(((((.(((((........))))))))))...).))))))).(....)))). ( -29.30, z-score = -2.50, R) >droSim1.chrX 9559445 98 - 17042790 --CACCCAU-CCAC--UCGAUAAAGUGCCACCCGCUCAACUUCUGCUCCCGUGAGCAGAACAAUUUCCAAUGUUCGCUCAACUGUGGUGGCAACGAAAGGAGC --......(-((..--(((......((((((((((.........)).....(((((.(((((........))))))))))...).))))))).)))..))).. ( -29.80, z-score = -2.67, R) >droGri2.scaffold_14853 2585017 90 + 10151454 ----------CCUUCCCCACUUUUGGGCUGC---CUCAACUUCUGCUCCCGCAAGCAGAACAAUUUCCAAUGUGCGCUCAACUGUAGUGGCAAACAAAGGGGU ----------....((((((..((((((.((---.......((((((......))))))(((........)))))))))))..)).((.....))...)))). ( -24.70, z-score = -0.44, R) >consensus __CACCCAC_CCAC__UCGAUAAAGUGCCACCCGCUCAACUUCUGCUCCCGUGAGCAGAACAAUUUCCAAUGUUCGCUCAACUGUGGUGGCAACGAAAGGAGC ..........................((((((........((((((((....))))))))...............((......))))))))............ (-17.71 = -17.22 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:34 2011