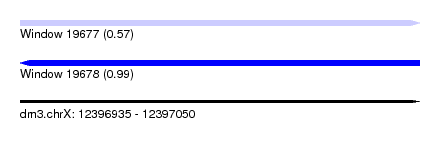
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,396,935 – 12,397,050 |
| Length | 115 |
| Max. P | 0.987874 |

| Location | 12,396,935 – 12,397,050 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.46433 |
| G+C content | 0.41339 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
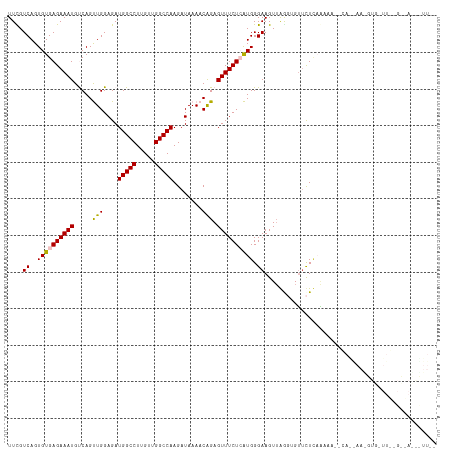
>dm3.chrX 12396935 115 + 22422827 UUCGUCAGUGUGAGAAAUGCCAGUUGGAGAUGGCCUUGUUGGCCAAGAUAAAACAGAGUUUCUCAUGGGAAGUUAAGUGUUCUAAACAAACAAAAAAAGUGUUUUUGCGAUAUUU-- .(((.(((.(((((((((....(((..(..(((((.....)))))...)..)))...))))))))).....(((.((....)).))).................)))))).....-- ( -24.40, z-score = -0.40, R) >droEre2.scaffold_4690 4286756 114 - 18748788 UUUGUCAGUGUGAGAAAUGUCAGCUGGAGAUGGCCUUGUUGGCCAAGAUAAA-CAGAGUUUCUGGUGGGAAGUUGAGAGUGCUCAAAAA--CAGGAAGGUGAUUGAGCUACGCUUAA ......((((((.....(.((((((.....(((((.....))))).......-(((.....)))......)))))).)..((((((..(--(......))..))))))))))))... ( -31.70, z-score = -1.14, R) >droYak2.chrX 6672880 115 + 21770863 UUUGUCAGUGUGAGAAAUGUCAGCUGGAGAUGGCCUUGUUGGCCAAGAUAAAACGGAGUUUCUGGUGGGAAGUAGGGAGGUCUCACAAA--CAGGAAGGUGUUUGAGUUAUGUUUAC .((((...(((((((..(.((.(((.....(((((.....)))))........(((.....)))......))).)).)..))))))).)--)))(((.(((.......))).))).. ( -28.40, z-score = -0.55, R) >droSec1.super_21 153375 92 + 1102487 UUCGUCAGUGUGAGAAAUGUCAGUUGGAGAUGGCCUUGUUGGCCAAGAUAAAACAGAGUUUCUCAUGGGAAGUUAGUUGUUUUUCGAUAUUU------------------------- ...(((...(((((((((.((.(((..(..(((((.....)))))...)..))).))))))))))).(((((........))))))))....------------------------- ( -23.60, z-score = -1.78, R) >droSim1.chrX_random 3372263 92 + 5698898 UUCGUCAGUGUGAGAAAUGCCAGUUGGAGAUGGCCUUGUUGGCCAAGAUAAAACAGAGUUUCUCAUGGGAAGUUAGUUGUUUUUCGAUAUUU------------------------- ...(((...(((((((((....(((..(..(((((.....)))))...)..)))...))))))))).(((((........))))))))....------------------------- ( -23.20, z-score = -1.38, R) >consensus UUCGUCAGUGUGAGAAAUGUCAGUUGGAGAUGGCCUUGUUGGCCAAGAUAAAACAGAGUUUCUCAUGGGAAGUUAGGUGUUCUCAAAAA__CA__AA_GUG_UU__G__A___UU__ ....((..((((((((((............(((((.....)))))............)))))))))).))............................................... (-16.31 = -16.47 + 0.16)

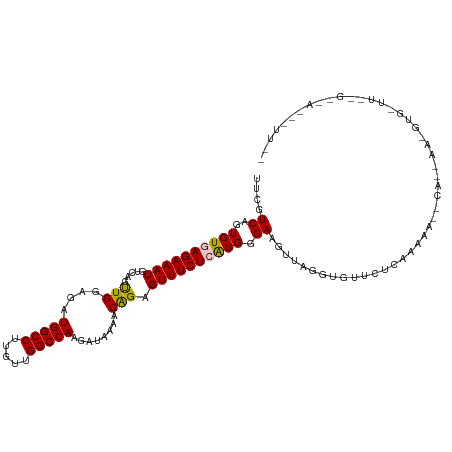
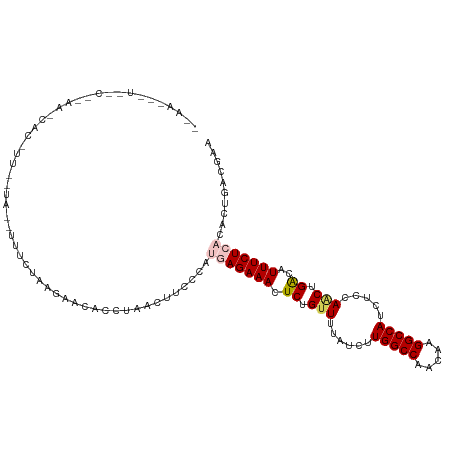
| Location | 12,396,935 – 12,397,050 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.46433 |
| G+C content | 0.41339 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12396935 115 - 22422827 --AAAUAUCGCAAAAACACUUUUUUUGUUUGUUUAGAACACUUAACUUCCCAUGAGAAACUCUGUUUUAUCUUGGCCAACAAGGCCAUCUCCAACUGGCAUUUCUCACACUGACGAA --.....(((((.............(((((.....)))))............(((((((..(((((......(((((.....))))).....))).))..)))))))...)).))). ( -21.20, z-score = -1.80, R) >droEre2.scaffold_4690 4286756 114 + 18748788 UUAAGCGUAGCUCAAUCACCUUCCUG--UUUUUGAGCACUCUCAACUUCCCACCAGAAACUCUG-UUUAUCUUGGCCAACAAGGCCAUCUCCAGCUGACAUUUCUCACACUGACAAA ...((.((.((((((..((......)--)..)))))).................(((((.((.(-((.....(((((.....))))).....))).))..))))).)).))...... ( -22.20, z-score = -1.81, R) >droYak2.chrX 6672880 115 - 21770863 GUAAACAUAACUCAAACACCUUCCUG--UUUGUGAGACCUCCCUACUUCCCACCAGAAACUCCGUUUUAUCUUGGCCAACAAGGCCAUCUCCAGCUGACAUUUCUCACACUGACAAA ........................((--(((((((((.........(((......)))..((.(((......(((((.....))))).....))).))....)))))))..)))).. ( -19.80, z-score = -1.80, R) >droSec1.super_21 153375 92 - 1102487 -------------------------AAAUAUCGAAAAACAACUAACUUCCCAUGAGAAACUCUGUUUUAUCUUGGCCAACAAGGCCAUCUCCAACUGACAUUUCUCACACUGACGAA -------------------------...........................(((((((.((.(((......(((((.....))))).....))).))..))))))).......... ( -16.70, z-score = -2.87, R) >droSim1.chrX_random 3372263 92 - 5698898 -------------------------AAAUAUCGAAAAACAACUAACUUCCCAUGAGAAACUCUGUUUUAUCUUGGCCAACAAGGCCAUCUCCAACUGGCAUUUCUCACACUGACGAA -------------------------...........................(((((((..(((((......(((((.....))))).....))).))..))))))).......... ( -16.40, z-score = -2.04, R) >consensus __AA___U__C__AA_CAC_UU__UA__UUUCUAAGAACACCUAACUUCCCAUGAGAAACUCUGUUUUAUCUUGGCCAACAAGGCCAUCUCCAACUGACAUUUCUCACACUGACGAA ....................................................(((((((.((.(((......(((((.....))))).....))).))..))))))).......... (-12.96 = -13.60 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:29 2011