
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,388,545 – 12,388,663 |
| Length | 118 |
| Max. P | 0.567927 |

| Location | 12,388,545 – 12,388,663 |
|---|---|
| Length | 118 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 61.97 |
| Shannon entropy | 0.78098 |
| G+C content | 0.58815 |
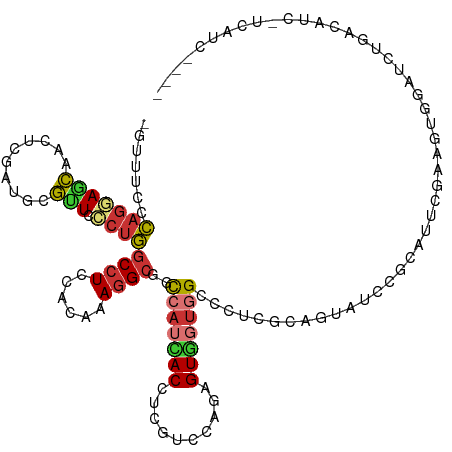
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.46 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
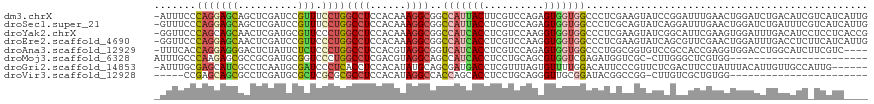
>dm3.chrX 12388545 118 + 22422827 -AUUUCCCAGGAGCAGCUCGAUCCGUUUCCUGGCCUCCACAAAGGCGGCCAUUACUUCGUCCAGAGUGGUGGCCCUCGAAGUAUCCGGAUUUGAACUGGAUCUGACAUCGUCAUCAUUG -......((((..(((.((((((((..(.((.((((......))))(((((((((((......))))))))))).....)).)..)))).)))).)))..))))............... ( -37.80, z-score = -1.22, R) >droSec1.super_21 145190 118 + 1102487 -GUUUCCCAGGAGCAGCUCGAUCCGUUUCCUGGCCUCCACAAAGGCGGCCAUUACCUCGUCCAGAGUGGUGGCCCUCGCAGUAUCAGGAUUUGAACUGGAUCUGAUUUCGUCAUCAUUG -((((.....)))).....(((((((((((((((((......))))(((((((((.((.....)))))))))))..........)))))....))).)))))((((......))))... ( -38.10, z-score = -1.24, R) >droYak2.chrX 6663908 118 + 21770863 -GGUUCCCAGCAGCAACUCGAUGCGUUCCCUGGCCUCCACAAAGGCGGCCAUCACCUCGUCCAAGGUGGUGGCCCUCGAAGUAUCGGCAUUCGAAGUGGAUUUGACAUCCUCCUCACCG -(((........((....((((((.(((....((((......))))(((((((((((......)))))))))))...)))))))))))....((.(.((((.....)))).).))))). ( -42.50, z-score = -2.17, R) >droEre2.scaffold_4690 4278266 118 - 18748788 -GGUUCCCAGGAGCAACUCGAUCCGUUCCCUGGCCUCCACAAAGGCGGCCAUCACCUCGUCCAAGGUGGUGGCCCUCGAAGUAUCAGCGUUCGAACUGGAUUUGACCUCUUCAUCAUUG -.((((....))))...(((((((((((((((((((......))))(((((((((((......)))))))))))..........))).)...)))).))).)))).............. ( -40.00, z-score = -1.78, R) >droAna3.scaffold_12929 663756 114 - 3277472 -UUUCACCAGGAGGGACUCUAUUCUCUCCCUGGCCUCCACGUAGGCGGUCAUCACCUCGUCCAGAGUGGUGGCCCUGGCGGUGUCCGCCACCGAGGUGGACCUGGCAUCUUCGUC---- -.....((((((((((......))))).))))).....(((.(((.(((((((((.((.....))))))))))).((.(((.(((((((.....)))))))))).))))).))).---- ( -49.30, z-score = -1.59, R) >droMoj3.scaffold_6328 1897866 95 + 4453435 AUUUGCCCAAGAGCGCCGCGAUGCGGUCCCUGGCCUCGACGUAGGCAGCCAUCACCUCCUGCAGCGUGGUCGAGAUGGUCGC-CUUGGGCUCGUGG----------------------- ....(((((((.((((((((.((.(((.....))).)).))).))).((((((.(..((........))..).)))))).))-)))))))......----------------------- ( -40.70, z-score = -0.87, R) >droGri2.scaffold_14853 2537413 112 - 10151454 -AUUUGCGAGCAUCGCCUCAAUGCGAUCCCUCACCUCCACAUAUGCAGCGAUGACCUCGUUUAGUGUUUUGGACAUUCCCGUUCUCGACUUCCUAUUUACAUUGUUGCCAUUG------ -......(((.(((((......)))))..)))............(((((((((........(((.(((..((((......))))..)))...)))....))))))))).....------ ( -23.30, z-score = -1.66, R) >droVir3.scaffold_12928 918681 90 + 7717345 -----CCGAGCAGCGCCUCGAUGCGCUCGCGCGCCUCCACAUAGGCCACCAGCACCUCCUGCAGGGUUGCGGAUACGGCCGG-CUUGUCGCUGUGG----------------------- -----.((.(((((((......))))).)).))...((((...((((.((.((((((......))).)))))....))))((-(.....)))))))----------------------- ( -32.80, z-score = 1.35, R) >consensus _GUUUCCCAGGAGCAACUCGAUGCGUUCCCUGGCCUCCACAAAGGCGGCCAUCACCUCGUCCAGAGUGGUGGCCCUCGCAGUAUCCGCAUUCGAAGUGGAUCUGACAUC_UCAUC____ .......(((((((..........))).))))((((......))))..(((((((..........)))))))............................................... (-12.32 = -12.46 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:27 2011