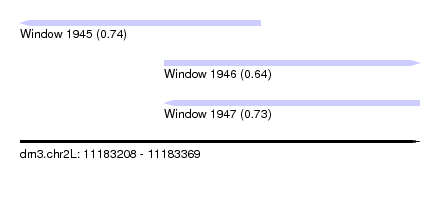
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,183,208 – 11,183,369 |
| Length | 161 |
| Max. P | 0.740248 |

| Location | 11,183,208 – 11,183,305 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.61 |
| Shannon entropy | 0.18121 |
| G+C content | 0.41984 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11183208 97 - 23011544 GC-AAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGCUUUACCUUUUCAUGUUGUGUCAUCAUCAGC-U-UCAAC---------- ((-....(((((((..(((((....))))).)))))))................(((((((...........)))))))...........))-.-.....---------- ( -19.50, z-score = -1.30, R) >droSim1.chr2L 10982923 97 - 22036055 GC-AAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGCUUUACCUUUUCAUGUUGUGUCAUCAUCAGC-U-UCAAC---------- ((-....(((((((..(((((....))))).)))))))................(((((((...........)))))))...........))-.-.....---------- ( -19.50, z-score = -1.30, R) >droSec1.super_3 6584380 97 - 7220098 GC-AAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGCUUUACCUUUUCAUGUUGUGUCAUCAUCAGC-U-UCAAC---------- ((-....(((((((..(((((....))))).)))))))................(((((((...........)))))))...........))-.-.....---------- ( -19.50, z-score = -1.30, R) >droYak2.chr2L 7581365 98 - 22324452 GC-AAAUUUACGUCCCCUGUCAAGAGGCAGCGUCGUAGCGUAUUUAAUAUUUUUCAGCGUGCUUUACCUUUUCAUGUUGUGUCAUCAUCAGCAU-ACAGC---------- ((-.....((((.(..(((((....))))).).))))((((...............))))))...........((((((((....)).))))))-.....---------- ( -17.16, z-score = -0.28, R) >droAna3.scaffold_12916 11912477 99 + 16180835 GC-AAAUUUACGUCCGCUGUCAAGAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCUUUUCAUGUUGUGUCAUCAUCAGCAUCUCGGC---------- ((-....(((((((.((((((....)))))))))))))................(((((((...........)))))))...........))........---------- ( -25.50, z-score = -2.32, R) >dp4.chr4_group3 6550285 98 - 11692001 GC-AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCUUUUCAUGUUGUGUCAUCAUCAGCAU-CAGGC---------- ((-....(((((((.((((((....)))))))))))))................(((((((...........)))))))...........))..-.....---------- ( -25.50, z-score = -2.88, R) >droPer1.super_1 8041644 98 - 10282868 GC-AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCUUUUCAUGUUGUGUCAUCAUCAGCAU-CAGGC---------- ((-....(((((((.((((((....)))))))))))))................(((((((...........)))))))...........))..-.....---------- ( -25.50, z-score = -2.88, R) >droWil1.scaffold_180708 7960129 97 + 12563649 GC-AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCUUUUCAUGUUGUGUCAUCAUCAGCA--UCAGC---------- ((-....(((((((.((((((....)))))))))))))................(((((((...........)))))))...........)).--.....---------- ( -25.50, z-score = -3.27, R) >droVir3.scaffold_12963 17889891 107 + 20206255 GC-AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCAUUUCAUGUUGUGUCAUCAUCAUCG--UCAGCAUCAGCACCA ((-.....((((((.((((((....))))))))))))((...............(((((((...........)))))))((.(.........)--.))))....)).... ( -26.20, z-score = -3.27, R) >droMoj3.scaffold_6500 4796038 107 + 32352404 GC-AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCAUUUCAUGUUGUGUCAUCAUCAUCG--UCAGCAUCAGCAGCG ((-.....((((((.((((((....))))))))))))(((..............(((((((...........)))))))............))--)..((....)).)). ( -26.97, z-score = -2.99, R) >droGri2.scaffold_15126 6966248 98 + 8399593 GCAAAAUUUACGUUCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCAUUUCAUGUUGUGUCAUCAUCAUUG--UCAGC---------- ........(((((.(((((((....))))))))))))((...............(((((((...........)))))))((.((.......))--.))))---------- ( -24.30, z-score = -2.63, R) >consensus GC_AAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCGUAUUUAAUAUUUUUCAGCGUGUUUUACCUUUUCAUGUUGUGUCAUCAUCAGCAU_UCAGC__________ .......(((((((..(((((....))))).)))))))................(((((((...........)))))))............................... (-16.68 = -16.69 + 0.01)


| Location | 11,183,266 – 11,183,369 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 67.77 |
| Shannon entropy | 0.67479 |
| G+C content | 0.60410 |
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -8.97 |
| Energy contribution | -9.08 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11183266 103 + 23011544 CGCUACGUCGCUGCCUCUUGACAGGGGACGUAAAUUUGCUUUUGCCAGCGCACCC----CUCUUAUCCCCCACACAACCAACGCCCCAGCCCCUGCCCCUUCAUCUG .((...((((((((((((....)))))..(((((......)))))))))).))..----.......................((....))....))........... ( -15.90, z-score = 0.82, R) >droSim1.chr2L 10982981 103 + 22036055 CGCUACGUCGCUGCCUCUUGACAGGGGACGUAAAUUUGCUUUUGCCAGCGCACCC----CUCUUAUCCCCCACACACCCAACGCCCCUGCCCCUCUACCUUCUCCUG ......((((((((((((....)))))..(((((......)))))))))).))..----................................................ ( -14.90, z-score = 0.39, R) >droSec1.super_3 6584438 103 + 7220098 CGCUACGUCGCUGCCUCUUGACAGGGGACGUAAAUUUGCUUUUGCCAGCGCACCC----CUCUUAUCCUCCACACACCCAACGCCCCUGUCCCUCUACCUUCUUCUG .((......))........((((((((.(((......(((......)))(.....----)....................)))))))))))................ ( -19.00, z-score = -1.30, R) >droEre2.scaffold_4929 12382052 101 - 26641161 CGCUACGUCGCUGCCUCUUGACAGGGGACGUAAAUUUGCUUUUGCCAGCGCACCC----CUCUGAUGCCCCACACAGCCAACGCCCCUGCCCUUU--CUGUCUCCUG .((......))..........(((((((((((((......)))))..(((.....----..(((.((.....)))))....)))...........--..)))))))) ( -19.20, z-score = 0.98, R) >droAna3.scaffold_12916 11912537 89 - 16180835 CGCUACGUCGCUGCCUCUUGACAGCGGACGUAAAUUUGCUUUUGCCUCCGCUCCU---UGCACCGUGGCCCGUGG-ACC-CCUACCCUGUCUCA------------- .(((((((((........))))((((((.(((((......))))).))))))...---......)))))..((((-...-.)))).........------------- ( -25.50, z-score = -1.99, R) >dp4.chr4_group3 6550344 83 + 11692001 CGCUACGUCGCUGCCUUUUGACAGCGGACGUAAAUUUGCUUUUGCCUGCUCAGCC---CCUGCCCCUGCCC-UGC-CCC-CCUACCCCC------------------ .((((((((((((.(....).)))).)))))).....((....))..))...((.---...((....))..-.))-...-.........------------------ ( -15.50, z-score = -1.26, R) >droPer1.super_1 8041703 78 + 10282868 CGCUACGUCGCUGCCUUUUGACAGCGGACGUAAAUUUGCUUUUGCCUGCUCAGCC---CCUGCCCCUGCCC-CAC-UCC-CCUC----------------------- .((((((((((((.(....).)))).)))))).....((........))...)).---.............-...-...-....----------------------- ( -15.40, z-score = -1.02, R) >droWil1.scaffold_180708 7960187 95 - 12563649 CGCUACGUCGCUGCCUUUUGACAGCGGACGUAAAUUUGCUUUUGCCUGCCUCCCCCCGUGUCCCUUAGCCACCACUCCC-CCUCCCCCACAUCACG----------- .((((((((((((.(....).)))).)))))).....((....))..)).......((((...................-............))))----------- ( -15.58, z-score = -1.49, R) >droVir3.scaffold_12963 17889959 84 - 20206255 CGCUACGUCGCUGCCUUUUGACAGCGGACGUAAAUUUGCUUUUGCC--CAUGCCC----UUGCCCCCACCCAACCAACCGCAAACCUGGG----------------- ...((((((((((.(....).)))).))))))............((--(((((..----(((............)))..)))....))))----------------- ( -18.00, z-score = -0.38, R) >droMoj3.scaffold_6500 4796106 104 - 32352404 CGCUACGUCGCUGCCUUUUGACAGCGGACGUAAAUUUGCUUUUGCCCGUGCCCCU---UGCCUCGUACCCCCUCCUUCCAUCAGUCCCGUCCCUCUGCCACUCUGCC .((......((((.(....).))))(((((((((......)))))..((((....---......))))....................))))....))......... ( -16.40, z-score = -0.28, R) >consensus CGCUACGUCGCUGCCUCUUGACAGCGGACGUAAAUUUGCUUUUGCCAGCGCACCC____CUCCCAUCCCCCACACACCCACCGCCCCUGCCCCU_____________ .((((((((.(((.(....).)))..)))))).....((....))..)).......................................................... ( -8.97 = -9.08 + 0.11)


| Location | 11,183,266 – 11,183,369 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.77 |
| Shannon entropy | 0.67479 |
| G+C content | 0.60410 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11183266 103 - 23011544 CAGAUGAAGGGGCAGGGGCUGGGGCGUUGGUUGUGUGGGGGAUAAGAG----GGGUGCGCUGGCAAAAGCAAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCG .................((((.(.(((((.((.((((((((((...((----(..(((..........)))..)))..)))))))).))..)).))))).).)))). ( -31.40, z-score = -0.21, R) >droSim1.chr2L 10982981 103 - 22036055 CAGGAGAAGGUAGAGGGGCAGGGGCGUUGGGUGUGUGGGGGAUAAGAG----GGGUGCGCUGGCAAAAGCAAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCG .........((.....(((((((((((...((.(((.(((.((.....----..)).).)).)))...))......))).)))))))).....)).((......)). ( -29.10, z-score = -0.48, R) >droSec1.super_3 6584438 103 - 7220098 CAGAAGAAGGUAGAGGGACAGGGGCGUUGGGUGUGUGGAGGAUAAGAG----GGGUGCGCUGGCAAAAGCAAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCG .........((.....(((((((((((..((((..(............----..)..)))).((....))......))).)))))))).....)).((......)). ( -27.94, z-score = -0.60, R) >droEre2.scaffold_4929 12382052 101 + 26641161 CAGGAGACAG--AAAGGGCAGGGGCGUUGGCUGUGUGGGGCAUCAGAG----GGGUGCGCUGGCAAAAGCAAAUUUACGUCCCCUGUCAAGAGGCAGCGACGUAGCG ..(....)..--....(((((((((((..(((.(((.(((((((....----.))))).)).)))..)))......))).))))))))........((......)). ( -37.70, z-score = -1.95, R) >droAna3.scaffold_12916 11912537 89 + 16180835 -------------UGAGACAGGGUAGG-GGU-CCACGGGCCACGGUGCA---AGGAGCGGAGGCAAAAGCAAAUUUACGUCCGCUGUCAAGAGGCAGCGACGUAGCG -------------.....(...(((..-(((-(....))))....))).---..).((....((....)).....((((((.((((((....)))))))))))))). ( -28.00, z-score = -0.91, R) >dp4.chr4_group3 6550344 83 - 11692001 ------------------GGGGGUAGG-GGG-GCA-GGGCAGGGGCAGG---GGCUGAGCAGGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG ------------------(((.(((((-...-((.-..((..(..(((.---..)))..)..))....))...))))).)))((((((....))))))......... ( -25.70, z-score = -1.81, R) >droPer1.super_1 8041703 78 - 10282868 -----------------------GAGG-GGA-GUG-GGGCAGGGGCAGG---GGCUGAGCAGGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG -----------------------....-...-...-..((....((...---.(((.....)))....)).....((((((.((((((....)))))))))))))). ( -24.10, z-score = -1.34, R) >droWil1.scaffold_180708 7960187 95 + 12563649 -----------CGUGAUGUGGGGGAGG-GGGAGUGGUGGCUAAGGGACACGGGGGGAGGCAGGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG -----------(((.((((...(((.(-..((...((.(((......(......)......)))....))...))..).)))((((((....)))))).)))).))) ( -23.70, z-score = -0.28, R) >droVir3.scaffold_12963 17889959 84 + 20206255 -----------------CCCAGGUUUGCGGUUGGUUGGGUGGGGGCAA----GGGCAUG--GGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG -----------------(((..((((.((.((....)).)).))))..----)))....--.((....))....(((((((.((((((....))))))))))))).. ( -27.00, z-score = -0.44, R) >droMoj3.scaffold_6500 4796106 104 + 32352404 GGCAGAGUGGCAGAGGGACGGGACUGAUGGAAGGAGGGGGUACGAGGCA---AGGGGCACGGGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG .((....((((((..(((((...((......))......(..((..((.---....)).))..).............))))).))))))....)).((......)). ( -27.40, z-score = -0.92, R) >consensus _____________AGGGGCAGGGGAGGUGGGUGUGUGGGGGAUGAGAG____GGGUGCGCAGGCAAAAGCAAAUUUACGUCCGCUGUCAAAAGGCAGCGACGUAGCG ..........................................................((..((....)).....((((((..(((((....))))).)))))))). (-13.07 = -13.37 + 0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:44 2011