| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,361,196 – 12,361,305 |
| Length | 109 |
| Max. P | 0.861414 |

| Location | 12,361,196 – 12,361,305 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.46472 |
| G+C content | 0.37873 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
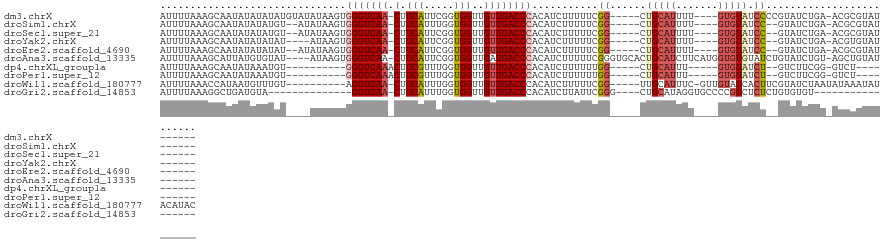
>dm3.chrX 12361196 109 + 22422827 AUUUUAAAGCAAUAUAUAUAUGUAUAUAAGUGGGUCAA-CUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGG-----CUGCAUUUU----GUGUAUCCCCGUAUCUGA-ACGCGUAU------ ........((..((((((....)))))).(((((((((-(.((((...))))..)))))))))).((.....(((-----.(((((...----)))))...))).....))-..))....------ ( -28.40, z-score = -2.32, R) >droSim1.chrX 9494065 105 + 17042790 AUUUUAAAGCAAUAUAUAUGU--AUAUAAGUGGGUCAA-CUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGG-----CUGCAUUUU----GUGUAUCC--GUAUCUGA-ACGCGUAU------ ...............((((((--......(((((((((-(.((((...))))..)))))))))).((.....(((-----.(((((...----))))).))--).....))-..))))))------ ( -30.00, z-score = -3.21, R) >droSec1.super_21 119081 105 + 1102487 AUUUUAAAGCAAUAUAUAUGU--AUAUAAGUGGGUCAA-CUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGG-----CUGCAUUUU----GUGUAUCC--GUAUCUGA-ACGCGUAU------ ...............((((((--......(((((((((-(.((((...))))..)))))))))).((.....(((-----.(((((...----))))).))--).....))-..))))))------ ( -30.00, z-score = -3.23, R) >droYak2.chrX 6636156 103 + 21770863 AUUUUAAAGCAAUAUAUAUAU----AUAAGUGGGUCAA-CUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGG-----CUGCAUUUU----GUGUAUCC--GUAUCUGA-ACGUGUAU------ ...........((((((....----....(((((((((-(.((((...))))..)))))))))).((.....(((-----.(((((...----))))).))--).....))-..))))))------ ( -28.60, z-score = -3.19, R) >droEre2.scaffold_4690 4252841 105 - 18748788 AUUUUAAAGCAAUAUAUAUAU--AUAUAAGUGGGUCAA-CUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGG-----CUGCAUUUU----GUGUAUCC--GUAUCUGA-ACGCGUAU------ ........((...........--......(((((((((-(.((((...))))..)))))))))).((.....(((-----.(((((...----))))).))--).....))-..))....------ ( -29.40, z-score = -3.27, R) >droAna3.scaffold_13335 2349449 114 - 3335858 AUUUUAAAGCAUUAUGUGUAU----AUAAGUGGGUCAA-CUUCAUUCGGUGGUUGAUGACCCACAUCUUUUUCGGGUGCACUGCAUCUUCAUGGUGUGUAUCUGUAUCUGU-AGCUGUAU------ .......(((...........----....((((((((.-(.((((...))))..).))))))))........((((((((.((((((.....))))))....)))))))).-.)))....------ ( -30.10, z-score = -1.45, R) >dp4.chrXL_group1a 3101528 93 + 9151740 AUUUUAAAGCAAUAUAAAUGU----------GGGUCAAACUUCGUUUGGUGGUUGUUGACCCACAUCUUUUUUGG-----CUGCAUUU-----GUGUAUCU--GUCUUCGG-GUCU---------- .......(((.......((((----------(((((((...(((.....)))...)))))))))))........)-----))......-----....((((--(....)))-))..---------- ( -22.86, z-score = -1.96, R) >droPer1.super_12 2237523 93 - 2414086 AUUUUAAAGCAAUAUAAAUGU----------GGGUCAAACUUCGUUUGGUGGUUGUUGACCCACAUCUUUUUUGG-----CUGCAUUU-----GUGUAUCU--GUCUUCGG-GUCU---------- .......(((.......((((----------(((((((...(((.....)))...)))))))))))........)-----))......-----....((((--(....)))-))..---------- ( -22.86, z-score = -1.96, R) >droWil1.scaffold_180777 4140986 109 + 4753960 AUUUUAAACCAUAAUGUUUGU----------AGGUCAA-CUUCAUUUGGUGGUUGUUGACCCACAUCUUUUUCGG-----UUGCAUUUC-GUUGUAUCACUUCGUAUCUAAUAUAAAUAUACAUAC .............((((((((----------(((((((-(.((((...))))..)))))))............((-----(.(((....-..)))))).............)))))))))...... ( -18.80, z-score = -0.86, R) >droGri2.scaffold_14853 2511602 90 - 10151454 AUUUUAAAGGCUGAUGUA--------------GGUCAA-CUUCAUUUGGUGGUUGUUGACCCACAUCUUAUUCGGG----CUGCAUAGGUGCCCCGUCUCUCUGUGUGU----------------- .......((((.(((((.--------------((((((-(.((((...))))..))))))).)))))......(((----(.((....)))))).))))..........----------------- ( -27.70, z-score = -1.90, R) >consensus AUUUUAAAGCAAUAUAUAUGU____AUAAGUGGGUCAA_CUUCAUUCGGUGGUUGUUGACCCACAUCUUUUUCGG_____CUGCAUUUU____GUGUAUCC__GUAUCUGA_ACGCGUAU______ ...............................(((((((...(((.....)))...)))))))................................................................ (-10.98 = -11.12 + 0.14)

| Location | 12,361,196 – 12,361,305 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.46472 |
| G+C content | 0.37873 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -7.91 |
| Energy contribution | -8.23 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
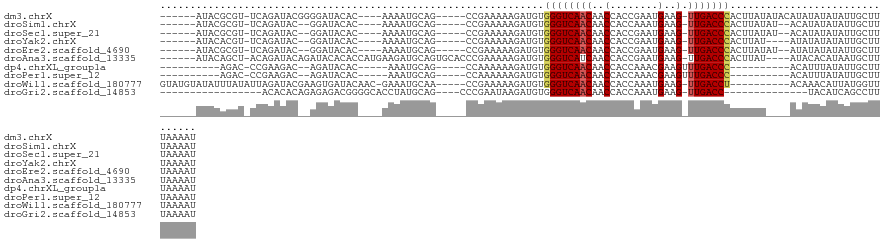
>dm3.chrX 12361196 109 - 22422827 ------AUACGCGU-UCAGAUACGGGGAUACAC----AAAAUGCAG-----CCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAG-UUGACCCACUUAUAUACAUAUAUAUAUUGCUUUAAAAU ------...(.(((-......))).).......----.....((((-----...........((((((((((..(........)..)-))))))))).((((((....))))))))))........ ( -24.00, z-score = -2.00, R) >droSim1.chrX 9494065 105 - 17042790 ------AUACGCGU-UCAGAUAC--GGAUACAC----AAAAUGCAG-----CCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAG-UUGACCCACUUAUAU--ACAUAUAUAUUGCUUUAAAAU ------....(((.-.......(--((...((.----....))...-----)))........((((((((((..(........)..)-)))))))))......--..........)))........ ( -21.60, z-score = -2.23, R) >droSec1.super_21 119081 105 - 1102487 ------AUACGCGU-UCAGAUAC--GGAUACAC----AAAAUGCAG-----CCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAG-UUGACCCACUUAUAU--ACAUAUAUAUUGCUUUAAAAU ------....(((.-.......(--((...((.----....))...-----)))........((((((((((..(........)..)-)))))))))......--..........)))........ ( -21.60, z-score = -2.07, R) >droYak2.chrX 6636156 103 - 21770863 ------AUACACGU-UCAGAUAC--GGAUACAC----AAAAUGCAG-----CCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAG-UUGACCCACUUAU----AUAUAUAUAUUGCUUUAAAAU ------.....(((-......))--).......----.....((((-----...........((((((((((..(........)..)-))))))))).(((----(....))))))))........ ( -20.10, z-score = -1.92, R) >droEre2.scaffold_4690 4252841 105 + 18748788 ------AUACGCGU-UCAGAUAC--GGAUACAC----AAAAUGCAG-----CCGAAAAAGAUGUGGGUCAACAACCACCGAAUGAAG-UUGACCCACUUAUAU--AUAUAUAUAUUGCUUUAAAAU ------....(((.-.......(--((...((.----....))...-----)))........((((((((((..(........)..)-)))))))))......--..........)))........ ( -21.60, z-score = -2.06, R) >droAna3.scaffold_13335 2349449 114 + 3335858 ------AUACAGCU-ACAGAUACAGAUACACACCAUGAAGAUGCAGUGCACCCGAAAAAGAUGUGGGUCAUCAACCACCGAAUGAAG-UUGACCCACUUAU----AUACACAUAAUGCUUUAAAAU ------.((.(((.-.............(((..(((....)))..)))..............(((((((((((.........)))..-.))))))))....----...........))).)).... ( -17.10, z-score = -0.23, R) >dp4.chrXL_group1a 3101528 93 - 9151740 ----------AGAC-CCGAAGAC--AGAUACAC-----AAAUGCAG-----CCAAAAAAGAUGUGGGUCAACAACCACCAAACGAAGUUUGACCC----------ACAUUUAUAUUGCUUUAAAAU ----------....-........--........-----......((-----(......((((((((((((((..(........)..).)))))))----------)))))).....)))....... ( -18.40, z-score = -2.94, R) >droPer1.super_12 2237523 93 + 2414086 ----------AGAC-CCGAAGAC--AGAUACAC-----AAAUGCAG-----CCAAAAAAGAUGUGGGUCAACAACCACCAAACGAAGUUUGACCC----------ACAUUUAUAUUGCUUUAAAAU ----------....-........--........-----......((-----(......((((((((((((((..(........)..).)))))))----------)))))).....)))....... ( -18.40, z-score = -2.94, R) >droWil1.scaffold_180777 4140986 109 - 4753960 GUAUGUAUAUUUAUAUUAGAUACGAAGUGAUACAAC-GAAAUGCAA-----CCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAG-UUGACCU----------ACAAACAUUAUGGUUUAAAAU ((.(((((.((..(((...)))..))...)))))))-.......((-----(((.......(((((((((((..(........)..)-)))))))----------))).......)))))...... ( -21.84, z-score = -2.05, R) >droGri2.scaffold_14853 2511602 90 + 10151454 -----------------ACACACAGAGAGACGGGGCACCUAUGCAG----CCCGAAUAAGAUGUGGGUCAACAACCACCAAAUGAAG-UUGACC--------------UACAUCAGCCUUUAAAAU -----------------........((((.(((((((....)))..----)))).....(((((((((((((..(........)..)-))))))--------------))))))...))))..... ( -29.80, z-score = -4.61, R) >consensus ______AUACACGU_UCAGAUAC__AGAUACAC____AAAAUGCAG_____CCGAAAAAGAUGUGGGUCAACAACCACCAAAUGAAG_UUGACCCACUUAU____ACAUAUAUAUUGCUUUAAAAU ................................................................(((((((....((.....))....)))))))............................... ( -7.91 = -8.23 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:24 2011