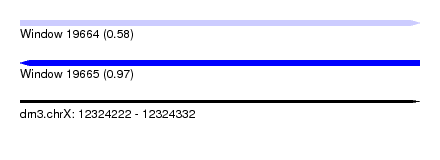
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,324,222 – 12,324,332 |
| Length | 110 |
| Max. P | 0.971723 |

| Location | 12,324,222 – 12,324,332 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.57009 |
| G+C content | 0.48033 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
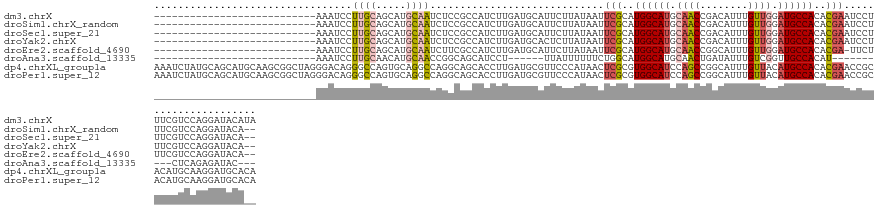
>dm3.chrX 12324222 110 + 22422827 ---------------------------AAAUCCUUGCAGCAUGCAAUCUCCGCCAUCUUGAUGCAUUCUUAUAAUUCGCAUGGCAUGCAACCGACAUUUGUUGGAUGCCACACGAAUCCUUUCGUCCAGGAUACAUA ---------------------------..(((((.((((.(((((.((...........))))))).))....(((((..((((((.((((........)))).))))))..)))))......))..)))))..... ( -28.80, z-score = -1.88, R) >droSim1.chrX_random 259012 108 + 5698898 ---------------------------AAAUCCUUGCAGCAUGCAAUCUCCGCCAUCUUGAUGCAUUCUUAUAAUUCGCAUGGCAUGCAACCGACAUUUGUUGGAUGCCACACGAAUCCUUUCGUCCAGGAUACA-- ---------------------------..(((((.((((.(((((.((...........))))))).))....(((((..((((((.((((........)))).))))))..)))))......))..)))))...-- ( -28.80, z-score = -1.91, R) >droSec1.super_21 83346 108 + 1102487 ---------------------------AAAUCCUUGCAGCAUGCAAUCUCCGCCAUCUUGAUGCAUUCUUAUAAUUCGCAUGGCAUGCAACCGACAUUUGUUGGAUGCCACACGAAUCCUUUCGUCCAGGAUACA-- ---------------------------..(((((.((((.(((((.((...........))))))).))....(((((..((((((.((((........)))).))))))..)))))......))..)))))...-- ( -28.80, z-score = -1.91, R) >droYak2.chrX 6601839 108 + 21770863 ---------------------------AAAUCCUUGCAGCAUGCAAUCUCCGCCAUCUUGAUGCACUCUUAUAAUUCGCAUGGCAUGCAACCGACAUUUGUUGGAUGCCACACGAAUCCUUUCGUCCAGGAUACA-- ---------------------------..(((((.((.((((.(((...........))))))).........(((((..((((((.((((........)))).))))))..)))))......))..)))))...-- ( -27.90, z-score = -1.76, R) >droEre2.scaffold_4690 4216650 107 - 18748788 ---------------------------AAAUCCUUGCAGCAUGCAAUCUUCGCCAUCUUGAUGCAUUCUUAUAAUUCGCAUGGCAUGCAACCGGCAUUUGUUGGAUGCCACACGA-UUCUUUCGUCCAGGAUACA-- ---------------------------..(((((.((((.(((((.((...........))))))).))......(((..((((((.((((........)))).))))))..)))-.......))..)))))...-- ( -27.20, z-score = -0.85, R) >droAna3.scaffold_13335 2308401 91 - 3335858 ---------------------------AAAUCCUUGCAACAUGCAACCGGCAGCAUCCU------UUAUUUUUUCUGGCAUGGCAUGCAACUGAUAUUUGUCGGUUGCCACAU----------CUCAGAGAUAC--- ---------------------------....((((((.....))))..)).........------......(((((((.(((....(((((((((....)))))))))..)))----------.)))))))...--- ( -23.80, z-score = -1.57, R) >dp4.chrXL_group1a 3059431 137 + 9151740 AAAUCUAUGCAGCAUGCAAGCGGCUAGGGACAGGGCCAGUGCAGGCCAGGCAGCACCUUGAUGCGUUCCCAUAACUCGCGUGGCAUCCAGCCGGCAUUUGUUACAUGCCACACGAACCGCACAUGCAAGGAUGCACA .......(((((((((...((((...(((((..((((......))))..(((.(.....).))))))))......(((.(((((((..(((........)))..))))))).))).)))).))))).....)))).. ( -46.90, z-score = 0.19, R) >droPer1.super_12 2194453 137 - 2414086 AAAUCUAUGCAGCAUGCAAGCGGCUAGGGACAGGGCCAGUGCAGGCCAGGCAGCACCUUGAUGCGUUCCCAUAACUCGCGUGGCAUCCAGCCGGCAUUUGUUACAUGCCACACGAACCGCACAUGCAAGGAUGCACA .......(((((((((...((((...(((((..((((......))))..(((.(.....).))))))))......(((.(((((((..(((........)))..))))))).))).)))).))))).....)))).. ( -46.90, z-score = 0.19, R) >consensus ___________________________AAAUCCUUGCAGCAUGCAAUCUCCGCCAUCUUGAUGCAUUCUUAUAAUUCGCAUGGCAUGCAACCGACAUUUGUUGGAUGCCACACGAAUCCUUUCGUCCAGGAUACA__ .................................((((.....)))).............................(((..((((((.((((........)))).))))))..)))...................... (-14.92 = -15.12 + 0.20)


| Location | 12,324,222 – 12,324,332 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.57009 |
| G+C content | 0.48033 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.47 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
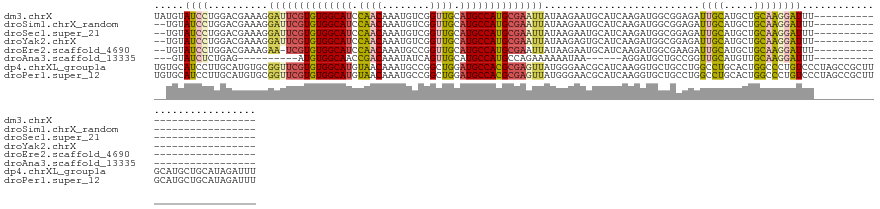
>dm3.chrX 12324222 110 - 22422827 UAUGUAUCCUGGACGAAAGGAUUCGUGUGGCAUCCAACAAAUGUCGGUUGCAUGCCAUGCGAAUUAUAAGAAUGCAUCAAGAUGGCGGAGAUUGCAUGCUGCAAGGAUUU--------------------------- .....(((((...(....)((((((((((((((.((((........)))).))))))))))))))...((.(((((((...........)).))))).))...)))))..--------------------------- ( -34.60, z-score = -1.65, R) >droSim1.chrX_random 259012 108 - 5698898 --UGUAUCCUGGACGAAAGGAUUCGUGUGGCAUCCAACAAAUGUCGGUUGCAUGCCAUGCGAAUUAUAAGAAUGCAUCAAGAUGGCGGAGAUUGCAUGCUGCAAGGAUUU--------------------------- --...(((((...(....)((((((((((((((.((((........)))).))))))))))))))...((.(((((((...........)).))))).))...)))))..--------------------------- ( -34.60, z-score = -1.68, R) >droSec1.super_21 83346 108 - 1102487 --UGUAUCCUGGACGAAAGGAUUCGUGUGGCAUCCAACAAAUGUCGGUUGCAUGCCAUGCGAAUUAUAAGAAUGCAUCAAGAUGGCGGAGAUUGCAUGCUGCAAGGAUUU--------------------------- --...(((((...(....)((((((((((((((.((((........)))).))))))))))))))...((.(((((((...........)).))))).))...)))))..--------------------------- ( -34.60, z-score = -1.68, R) >droYak2.chrX 6601839 108 - 21770863 --UGUAUCCUGGACGAAAGGAUUCGUGUGGCAUCCAACAAAUGUCGGUUGCAUGCCAUGCGAAUUAUAAGAGUGCAUCAAGAUGGCGGAGAUUGCAUGCUGCAAGGAUUU--------------------------- --...(((((...(....)((((((((((((((.((((........)))).))))))))))))))...((.(((((((...........)).))))).))...)))))..--------------------------- ( -34.90, z-score = -1.60, R) >droEre2.scaffold_4690 4216650 107 + 18748788 --UGUAUCCUGGACGAAAGAA-UCGUGUGGCAUCCAACAAAUGCCGGUUGCAUGCCAUGCGAAUUAUAAGAAUGCAUCAAGAUGGCGAAGAUUGCAUGCUGCAAGGAUUU--------------------------- --...(((((...(....)..-(((((((((((.((((........)))).)))))))))))..........((((.((.....((((...)))).)).)))))))))..--------------------------- ( -31.80, z-score = -1.21, R) >droAna3.scaffold_13335 2308401 91 + 3335858 ---GUAUCUCUGAG----------AUGUGGCAACCGACAAAUAUCAGUUGCAUGCCAUGCCAGAAAAAAUAA------AGGAUGCUGCCGGUUGCAUGUUGCAAGGAUUU--------------------------- ---(((((((((.(----------.(((((((..((((........))))..))))))))))))........------..)))))..((...(((.....))).))....--------------------------- ( -21.90, z-score = 0.07, R) >dp4.chrXL_group1a 3059431 137 - 9151740 UGUGCAUCCUUGCAUGUGCGGUUCGUGUGGCAUGUAACAAAUGCCGGCUGGAUGCCACGCGAGUUAUGGGAACGCAUCAAGGUGCUGCCUGGCCUGCACUGGCCCUGUCCCUAGCCGCUUGCAUGCUGCAUAGAUUU ((((((.....((((((((((((((((((((((.((.(........).)).))))))))))).....((((..((((....)))).....((((......))))...))))..)))))..))))))))))))..... ( -58.10, z-score = -1.54, R) >droPer1.super_12 2194453 137 + 2414086 UGUGCAUCCUUGCAUGUGCGGUUCGUGUGGCAUGUAACAAAUGCCGGCUGGAUGCCACGCGAGUUAUGGGAACGCAUCAAGGUGCUGCCUGGCCUGCACUGGCCCUGUCCCUAGCCGCUUGCAUGCUGCAUAGAUUU ((((((.....((((((((((((((((((((((.((.(........).)).))))))))))).....((((..((((....)))).....((((......))))...))))..)))))..))))))))))))..... ( -58.10, z-score = -1.54, R) >consensus __UGUAUCCUGGACGAAAGGAUUCGUGUGGCAUCCAACAAAUGUCGGUUGCAUGCCAUGCGAAUUAUAAGAAUGCAUCAAGAUGGCGGAGAUUGCAUGCUGCAAGGAUUU___________________________ .....((((..........((((((((((((((.((((........)))).))))))))))))))..........................((((.....))))))))............................. (-24.54 = -24.47 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:19 2011