| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,319,113 – 12,319,206 |
| Length | 93 |
| Max. P | 0.944448 |

| Location | 12,319,113 – 12,319,206 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Shannon entropy | -0.00000 |
| G+C content | 0.32258 |
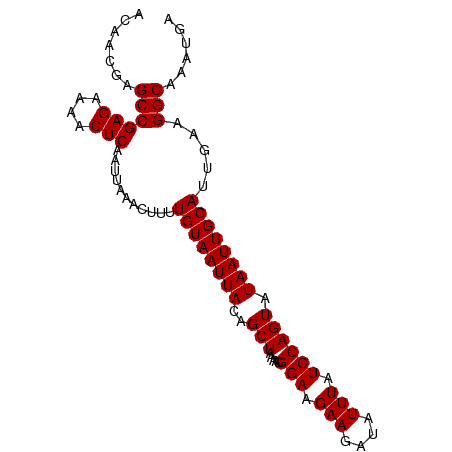
| Mean single sequence MFE | -16.20 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
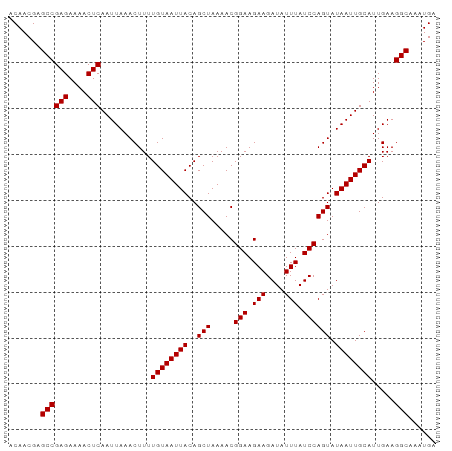
>dm3.chrX 12319113 93 + 22422827 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droGri2.scaffold_15081 471030 93 - 4274704 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droMoj3.scaffold_6359 526135 93 + 4525533 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droVir3.scaffold_12928 3938743 93 + 7717345 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droWil1.scaffold_180777 4075393 93 + 4753960 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droPer1.super_12 2186221 93 - 2414086 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >dp4.chrXL_group1a 3051147 93 + 9151740 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droEre2.scaffold_4690 4211506 93 - 18748788 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droYak2.chrX 6596574 93 + 21770863 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droSec1.super_21 78331 93 + 1102487 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >droSim1.chrX_random 254017 93 + 5698898 ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... ( -16.20, z-score = -1.80, R) >consensus ACAACGAGCCGAGAAAACUCAAUUAAACUUUUGUAAUUACAGCUAAAACGGAAGAAGAUAUUUAUCCAGUAUAAUUGCAUUGAAGGCAAAUGA .......((((((....)))...........((((((((..(((.....(((.(((....))).)))))).)))))))).....)))...... (-16.20 = -16.20 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:17 2011