
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,316,056 – 12,316,134 |
| Length | 78 |
| Max. P | 0.519200 |

| Location | 12,316,056 – 12,316,134 |
|---|---|
| Length | 78 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.04 |
| Shannon entropy | 0.47145 |
| G+C content | 0.44520 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -7.43 |
| Energy contribution | -7.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
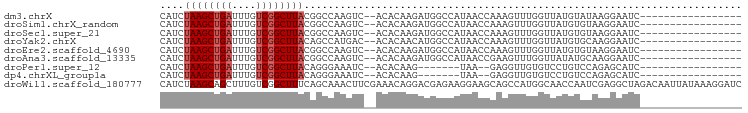
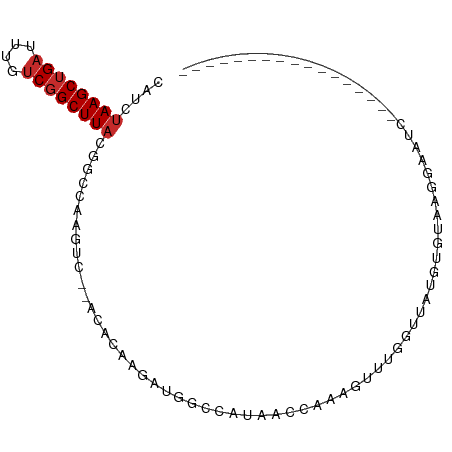
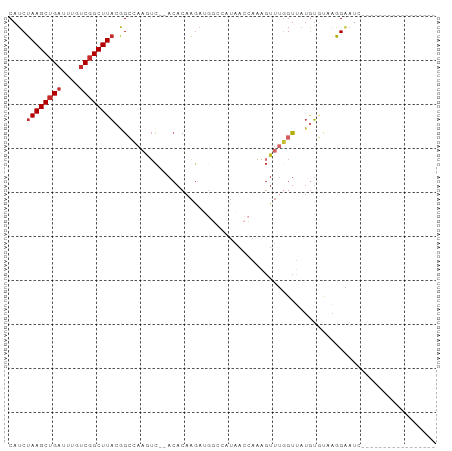
>dm3.chrX 12316056 78 + 22422827 CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC--ACACAAGAUGGCCAUAACCAAAGUUUGGUUAUGUAUAAGGAAUC----------------- ....((((((((....)))))))).(((((..((--......)))))))(((((((.....)))))))............----------------- ( -23.60, z-score = -3.10, R) >droSim1.chrX_random 251044 78 + 5698898 CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC--ACACAAGAUGGCCAUAACCAAAGUUUGGUUAUGUGUAAGGAAUC----------------- (((((....((((((((((.....))).))))))--)....)))))((((((((((.....)))))))).))........----------------- ( -24.50, z-score = -2.99, R) >droSec1.super_21 75364 78 + 1102487 CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC--ACACAAGAUGGCCAUAACCAAAGUUUGGUUAUGUGUAAGGAAUC----------------- (((((....((((((((((.....))).))))))--)....)))))((((((((((.....)))))))).))........----------------- ( -24.50, z-score = -2.99, R) >droYak2.chrX 6590666 78 + 21770863 CAUCUAAGCUGAUUUGUCGGCUUACAGCCAUGAC--ACACAACAUGGCCAUAACCAAAGUUUGGUUAUGUGCAAGGAAUC----------------- ....((((((((....))))))))..((((((..--......))))))((((((((.....))))))))...........----------------- ( -25.80, z-score = -3.37, R) >droEre2.scaffold_4690 4208284 78 - 18748788 CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC--ACACAAGAUGGCCAUAACCAAAGUUUGGUUAUGUGUAAGGAAUC----------------- (((((....((((((((((.....))).))))))--)....)))))((((((((((.....)))))))).))........----------------- ( -24.50, z-score = -2.99, R) >droAna3.scaffold_13335 1044573 78 + 3335858 CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC--ACACAAGAUGGCCAUAACCGAAGUUUGGUUAUAUGCAAGGAAUC----------------- ....((((((((....)))))))).(((((..((--......)))))))((((((((...))))))))............----------------- ( -23.30, z-score = -2.59, R) >droPer1.super_12 2181940 69 - 2414086 CAUCUAAGCUGAUUUGUCGGCUUACAGGGAAAUC--ACACAAG-------UAA--GAGGUUGUGUCCUGUCCAGAGCAUC----------------- ....((((((((....))))))))...(((....--((((((.-------(..--..).))))))....)))........----------------- ( -18.30, z-score = -1.24, R) >dp4.chrXL_group1a 3047045 69 + 9151740 CAUCUAAGCUGAUUUGUCGGCUUACAGGGAAAUC--ACACAAG-------UAA--GAGGUUGUGUCCUGUCCAGAGCAUC----------------- ....((((((((....))))))))...(((....--((((((.-------(..--..).))))))....)))........----------------- ( -18.30, z-score = -1.24, R) >droWil1.scaffold_180777 4072368 97 + 4753960 CAUCUAAGCAGCUUUGUCGGCUUUCAGCAAACUUCGAAACAGGACGAGAAGGAAGCAGCCAUGGCAACCAAUCGAGGCUAGACAAUUAUAAAGGAUC .((((........(((((((((((..((...((((............))))...)).((....))........)))))).))))).......)))). ( -19.26, z-score = 0.07, R) >consensus CAUCUAAGCUGAUUUGUCGGCUUACGGCCAAGUC__ACACAAGAUGGCCAUAACCAAAGUUUGGUUAUGUGUAAGGAAUC_________________ ....((((((((....))))))))......................................................................... ( -7.43 = -7.77 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:16 2011