| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,181,573 – 11,181,669 |
| Length | 96 |
| Max. P | 0.988073 |

| Location | 11,181,573 – 11,181,669 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Shannon entropy | 0.37156 |
| G+C content | 0.34360 |
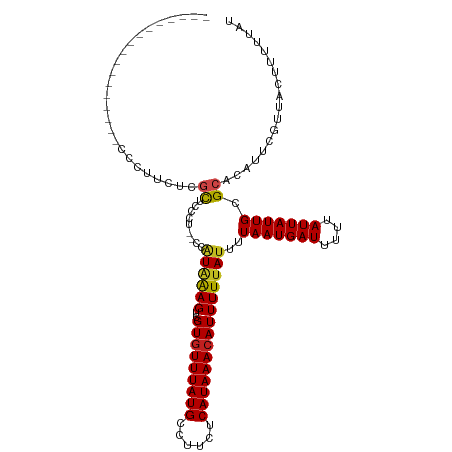
| Mean single sequence MFE | -13.44 |
| Consensus MFE | -10.11 |
| Energy contribution | -10.24 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.988073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
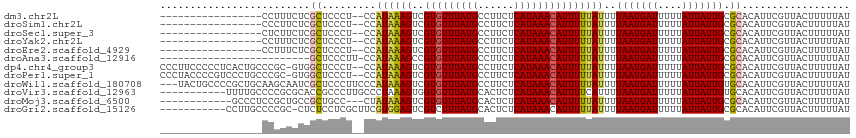
>dm3.chr2L 11181573 96 - 23011544 -----------------CCUUUCUCGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------------........((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -12.10, z-score = -3.11, R) >droSim1.chr2L 10981284 96 - 22036055 -----------------CCCUUCUCGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------------........((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -12.10, z-score = -3.20, R) >droSec1.super_3 6582736 96 - 7220098 -----------------CUCUUCUCGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------------........((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -12.10, z-score = -3.06, R) >droYak2.chr2L 7579813 96 - 22324452 -----------------CCUUUCUCGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------------........((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -12.10, z-score = -3.11, R) >droEre2.scaffold_4929 12380359 96 + 26641161 -----------------CCUUUCUCGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------------........((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -12.10, z-score = -3.11, R) >droAna3.scaffold_12916 11911130 89 + 16180835 -------------------------GCUCCCUU-CCAUAAAGCCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -------------------------((......-..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. ( -11.80, z-score = -2.57, R) >dp4.chr4_group3 6547233 112 - 11692001 CCCUUCCCCCUCACUGCCCGC-GUGGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU ..................((.-(((((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).))).))........... ( -15.30, z-score = -2.34, R) >droPer1.super_1 8038662 112 - 10282868 CCCUACCCCGUCCCUGCCCGC-GUGGCUCCCU--CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU ..................((.-(((((.....--..((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).))).))........... ( -15.30, z-score = -1.92, R) >droWil1.scaffold_180708 7958235 112 + 12563649 ---UACUGCCCCGCUGCAAGCAAUCGCUCCCUUCCCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGUGCACAUUCGUUACUUUUUAU ---.........(.(((((((....)))........((((((..(((((((((......)))))))))))))))..(((((((....))))))))))))................ ( -14.70, z-score = -1.51, R) >droVir3.scaffold_12963 17888741 104 + 20206255 -----------UUUUGCCCCGCGCACCGCCCUUGCCCUAAAGUGGUGUUUAUGCACUCUCAUAAACAUUUUCAUUUUAAUGAUUUUUAUUAUUGUGCACAUUCGUUACUUUUUAU -----------.........(((((..((....))..((((((((((((((((......)))))))))...)))))))..............))))).................. ( -17.70, z-score = -1.96, R) >droMoj3.scaffold_6500 4794858 100 + 32352404 ------------GCCCUCCGCUGCCGCUGCC---CUAUAAAGUCGUGUUUAUGCACUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU ------------.......(.(((.((....---..((((((..(((((((((......)))))))))))))))...((((((....))))))))))))................ ( -13.40, z-score = -1.05, R) >droGri2.scaffold_15126 6965053 103 + 8399593 -----------CCUUGCCCCGC-CUCUCCUCGCUUCGUGGAGUCGUCUUUAUGCACUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU -----------...(((...((-..((((.((...)).))))..))......(((.......(((((((........)))).))).......))))))................. ( -12.64, z-score = -0.90, R) >consensus _________________CCCUUCUCGCUCCCU__CCAUAAAGUCGUGUUUAUGCCUUCUCAUAAACAUUUUUAUUUUAAUGAUUUUUAUUAUUGCGCACAUUCGUUACUUUUUAU .........................((.........((((((..(((((((((......)))))))))))))))..(((((((....))))))).)).................. (-10.11 = -10.24 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:42 2011