| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,314,466 – 12,314,593 |
| Length | 127 |
| Max. P | 0.970224 |

| Location | 12,314,466 – 12,314,564 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Shannon entropy | 0.35471 |
| G+C content | 0.35453 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
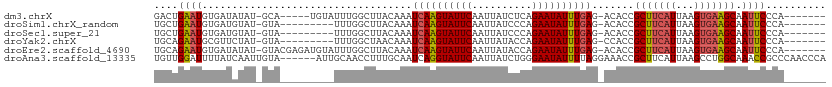
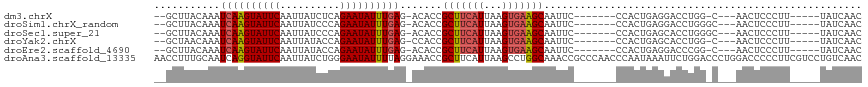
>dm3.chrX 12314466 98 + 22422827 GACUGAAUGUGAUAUAU-GCA-----UGUAUUUGGCUUACAAAUCAAGUAUUCAAUUAUCUCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUCCCA------- ....(((((((....((-((.-----((.(((((.....))))))).)))).......(((((((...))))))-)))).(((((((...))))))).))))...------- ( -22.10, z-score = -1.99, R) >droSim1.chrX_random 249472 94 + 5698898 UGCUGAAUGUGAUGUAU-GUA---------UUUGGCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUCCCA------- ....(((((((.((((.-((.---------....)).))))..((((((((((..........)))))))))).-.))).(((((((...))))))).))))...------- ( -22.60, z-score = -2.58, R) >droSec1.super_21 73797 94 + 1102487 UGCUGAAUGUGAUGUAU-GUA---------UUUGGCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUCCCA------- ....(((((((.((((.-((.---------....)).))))..((((((((((..........)))))))))).-.))).(((((((...))))))).))))...------- ( -22.60, z-score = -2.58, R) >droYak2.chrX 6589130 94 + 21770863 UGCAGAAUGCGUUCUAU-GUA---------UUUGGCUAACAAAUCAAGUAUUCAAUUAUACCAGAAUAUUUGAG-CCACCGCUUCAUUAAGUGAAGCAAUUCCCA------- .((.((((((((...))-)))---------))).)).......((((((((((..........)))))))))).-.....(((((((...)))))))........------- ( -23.70, z-score = -3.42, R) >droEre2.scaffold_4690 4206721 103 - 18748788 UGCAGAAUGUGAUAUAU-GUACGAGAUGUAUUUGGCUUACAAAUCAAGUAUUCAAUUAUACCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUCCCA------- ....(((((((......-........((((.......))))..((((((((((..........)))))))))).-.))).(((((((...))))))).))))...------- ( -21.60, z-score = -1.74, R) >droAna3.scaffold_13335 1042881 106 + 3335858 UGUUGGAUUUUAUCAAUUGUA------AUUGCAACCUUUGCAAUCAGGUAUUCAAUUAUCUGGGAAUAUUUUAGGAAACCGCUUCAUUAAGCCUGGCAAACCGCCCAACCCA .(((((...............------((((((.....))))))((((((......))))))((.........(....).((((....))))........))..)))))... ( -20.10, z-score = 0.40, R) >consensus UGCUGAAUGUGAUAUAU_GUA_________UUUGGCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG_ACACCGCUUCAUUAAGUGAAGCAAUUCCCA_______ ....((((...................................((((((((((..........)))))))))).......(((((((...))))))).)))).......... (-15.59 = -15.87 + 0.28)
| Location | 12,314,494 – 12,314,593 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.18 |
| Shannon entropy | 0.29328 |
| G+C content | 0.41327 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
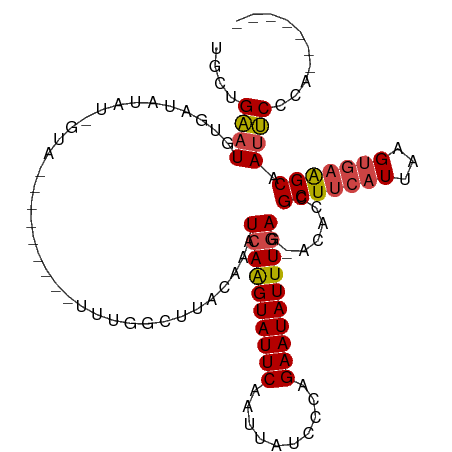
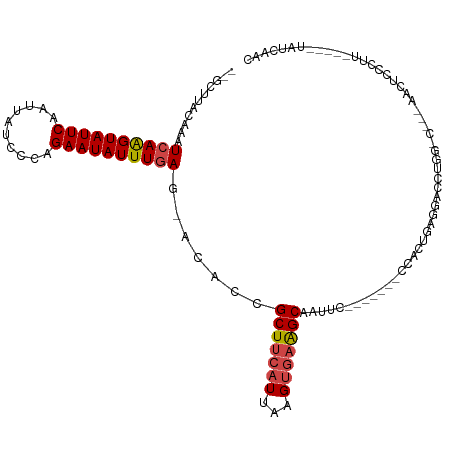
>dm3.chrX 12314494 99 + 22422827 --GCUUACAAAUCAAGUAUUCAAUUAUCUCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUC-------CCACUGAGGACCUGG-C---AACUCCCUU-----UAUCAAC --(((......((((((((((..........)))))))))).-.....(((((((...)))))))...((-------(......)))...))-)---.........-----....... ( -20.30, z-score = -2.45, R) >droSim1.chrX_random 249496 100 + 5698898 --GCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUC-------CCACUGAGGACCUGGGC---AACUCCCUU-----UAUCAAC --((.......((((((((((..........)))))))))).-...(((((((((...)))))))...((-------(......)))...))))---.........-----....... ( -22.20, z-score = -2.66, R) >droSec1.super_21 73821 100 + 1102487 --GCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUC-------CCACUGAGCACCUGGGC---AACUCCCUU-----UAUCAAC --(((((....((((((((((..........)))))))))).-.....(((((((...))))))).....-------....)))))....(((.---....)))..-----....... ( -24.50, z-score = -4.16, R) >droYak2.chrX 6589154 99 + 21770863 --GCUAACAAAUCAAGUAUUCAAUUAUACCAGAAUAUUUGAG-CCACCGCUUCAUUAAGUGAAGCAAUUC-------CCACUGAGCACCUGG-C---AACUCCCUU-----UAUCAAC --.........((((((((((..........))))))))))(-(((..(((((((...)))))))..(((-------.....)))....)))-)---.........-----....... ( -21.50, z-score = -3.81, R) >droEre2.scaffold_4690 4206754 99 - 18748788 --GCUUACAAAUCAAGUAUUCAAUUAUACCAGAAUAUUUGAG-ACACCGCUUCAUUAAGUGAAGCAAUUC-------CCACUGAGGACCCGG-C---AACUCCCUU-----UAUCAAC --(((......((((((((((..........)))))))))).-.....(((((((...)))))))...((-------(......)))...))-)---.........-----....... ( -20.40, z-score = -3.17, R) >droAna3.scaffold_13335 1042907 118 + 3335858 AACCUUUGCAAUCAGGUAUUCAAUUAUCUGGGAAUAUUUUAGGAAACCGCUUCAUUAAGCCUGGCAAACCGCCCAACCCAAUAAAUUCUGGACCCUGGACCCCCUUCGUCCUGUCAAC .....(((((..((((((......))))))(((((...((.((.....((((....))))..(((.....)))...)).))...)))))((((...((....))...)))))).))). ( -22.60, z-score = 0.32, R) >consensus __GCUUACAAAUCAAGUAUUCAAUUAUCCCAGAAUAUUUGAG_ACACCGCUUCAUUAAGUGAAGCAAUUC_______CCACUGAGGACCUGG_C___AACUCCCUU_____UAUCAAC ...........((((((((((..........)))))))))).......(((((((...)))))))..................................................... (-14.44 = -14.83 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:15 2011