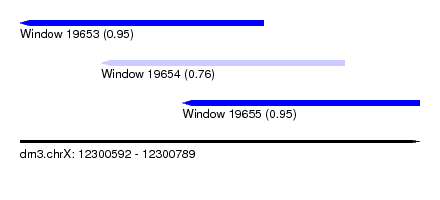
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,300,592 – 12,300,789 |
| Length | 197 |
| Max. P | 0.951097 |

| Location | 12,300,592 – 12,300,712 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.42178 |
| G+C content | 0.47917 |
| Mean single sequence MFE | -41.56 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.98 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12300592 120 - 22422827 CCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCCUUAUGCUAACGGCCAAUCAAGUUAAUGCACUUACUCAAAAAGGCAAGGAAAAGGUGGGCAUUACGUUUAUCGCCAGGAUGG (((((((((((((((((((......)))))))))))))...........(((............(((.(((.......)))))).......(((((((.....)))))))))).)))))) ( -39.80, z-score = -3.61, R) >droSim1.chrX 9459323 120 - 17042790 CCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUACUAACGGCCAAACAAGCUGAAGCACUCACUCAAAAAUGCAAUAAAAAGGUGGGCAUUCAGUUUAUCGCCAGGAUGG (((((((((((((((((((......)))))))))))))...........(((.....((((((((...((((((.................))))))..))))))))...))).)))))) ( -44.53, z-score = -6.27, R) >droSec1.super_21 56978 120 - 1102487 CCAUUCGGCCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUACUAACGGCCAAACAAGCUAAAGCACUCACUCAAAAAUGCAAUAAAAAUGUGGGCAUUCAGUUUAUCGCCAGGAUGG (((((((((((((((((((......)))))))))))))...........(((.((((..((....)).(((((...................)))))......))))...))).)))))) ( -38.11, z-score = -4.52, R) >droEre2.scaffold_4690 4194725 120 + 18748788 CCAUUCGACCCUUUGAUUUGAUCAAAAGUGGCAGGGUCAUUAUGCUAACGGCCAAUCAAGUGGAUGCACUUACUCAAAAGGGCAAGGAGAAGGUGGGCAUUCAGUUUGUGGCCAUGAUGG (((((.((((((.(.((((......)))).).))))))...........(((((...((.(((((((((((.(((...........))).))))..))))))).))..)))))..))))) ( -40.00, z-score = -1.96, R) >droYak2.chrX 6575873 120 - 21770863 CCACUCGACCCCAUGAUUUGAUCAAAAGUGGUAGGGUCAUUAUGCUAACCGCGAAUCAAGUGGAUGCAUUCACUCAAAGGGGCACGCAGAAUGUGCGCAUUCAGUUUGUGGCCAUGAUGG ((....(((((.((.((((......)))).)).)))))((((((....((((((((....(((((((.....((.....))(((((.....)))))))))))))))))))).)))))))) ( -38.80, z-score = -1.33, R) >droAna3.scaffold_13335 2282168 120 + 3335858 CCCGACGCCCUCUGAAGCUUCUGAGGAGCGGGCGUGCCAUUAUCCUGACCGCCGAUCAGGUGGAGGGGCUGACCCAGAACGGCAUGGAGAUGGUGGGCAUCCAGAUCUCCGUCAAGAUGG .(((.(((((((.((....)).)))).)))((((((((..........(((((.....))))).(((.....))).....)))))((((((..(((....))).)))))))))....))) ( -48.10, z-score = -0.41, R) >consensus CCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUGCUAACGGCCAAUCAAGUGGAUGCACUCACUCAAAAAGGCAAGGAAAAGGUGGGCAUUCAGUUUAUCGCCAGGAUGG (((((.((((((((.((((......)))).))))))))............(((......((....))...(((...................)))))).................))))) (-15.75 = -16.98 + 1.23)

| Location | 12,300,632 – 12,300,752 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.43197 |
| G+C content | 0.43889 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12300632 120 - 22422827 ACGAUAUCCUAUACCGGAAAACCAUUUGAUAUUAAGAAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCCUUAUGCUAACGGCCAAUCAAGUUAAUGCACUUACUCAAAAA ......(((......)))......(((((...((((..........(((((((((((((......)))))))))))))....(((((((..........))))..))))))).))))).. ( -28.60, z-score = -2.61, R) >droSim1.chrX 9459363 120 - 17042790 ACGAUAUCCUAUACUGGAAAACCCUUUGAUAUUAAGCAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUACUAACGGCCAAACAAGCUGAAGCACUCACUCAAAAA ......(((......)))......(((((......((.........(((((((((((((......)))))))))))))..........((((.......))))..))......))))).. ( -31.00, z-score = -3.47, R) >droSec1.super_21 57018 120 - 1102487 ACGAUAUCCUAUACUGGAAAACCCUUUGAUAUUAAGCAGUCCAUUCGGCCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUACUAACGGCCAAACAAGCUAAAGCACUCACUCAAAAA ......(((......)))......(((((......((.........(((((((((((((......)))))))))))))...........(((.......)))...))......))))).. ( -27.90, z-score = -2.38, R) >droEre2.scaffold_4690 4194765 120 + 18748788 ACAAUAUCCUAUACCGGAAGUGCCUACGAUAUUAAGAGGUCCAUUCGACCCUUUGAUUUGAUCAAAAGUGGCAGGGUCAUUAUGCUAACGGCCAAUCAAGUGGAUGCACUUACUCAAAAG ......(((......)))(((((((.(........)))(((((((.((((((.(.((((......)))).).)))))).....((.....))......)))))))))))).......... ( -27.50, z-score = -0.54, R) >droYak2.chrX 6575913 120 - 21770863 ACGAUAUCCUAUACCGGAAGUCCCUACGAUAUUAAGAAGUCCACUCGACCCCAUGAUUUGAUCAAAAGUGGUAGGGUCAUUAUGCUAACCGCGAAUCAAGUGGAUGCAUUCACUCAAAGG ..(((.(((......))).)))(((..((.........(((((((.(((((.((.((((......)))).)).)))))....(((.....))).....)))))))........))..))) ( -29.53, z-score = -1.12, R) >droAna3.scaffold_13335 2282208 120 + 3335858 AUCAUCUCCUACACGGGACAGGGCUACGAUUUGCGGAACACCCGACGCCCUCUGAAGCUUCUGAGGAGCGGGCGUGCCAUUAUCCUGACCGCCGAUCAGGUGGAGGGGCUGACCCAGAAC .(((.(((((...(((((...(((.(((.....(((.....))).(((((((.((....)).)))).)))..))))))....))))).(((((.....)))))))))).)))........ ( -46.70, z-score = -1.55, R) >consensus ACGAUAUCCUAUACCGGAAAACCCUACGAUAUUAAGAAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAUUAUGCUAACGGCCAAUCAAGUGGAUGCACUCACUCAAAAA ......(((......)))............................((((((((.((((......)))).))))))))....(((...((((.......))))..)))............ (-15.59 = -16.45 + 0.86)

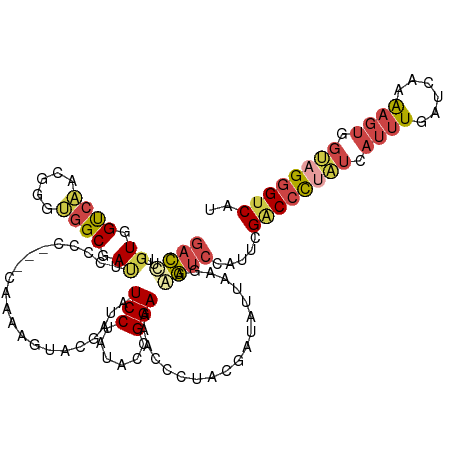
| Location | 12,300,672 – 12,300,789 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.50068 |
| G+C content | 0.47664 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12300672 117 - 22422827 GACCUUGUGGUCAACGGGUGGCGAUGUCCC---CAAAAGUACGAUAUCCUAUACCGGAAAACCAUUUGAUAUUAAGAAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCCU ((((....))))..(((((((.((((((..---.........)))))))))).)))...................(((.....)))(((((((((((((......))))))))))))).. ( -38.10, z-score = -2.79, R) >droSim1.chrX 9459403 117 - 17042790 GACUUUGUGGUCAACGGGUGGCGAUUAUCC---CAUAAGUACGAUAUCCUAUACUGGAAAACCCUUUGAUAUUAAGCAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAU ((((((...(((((.((((((.(.....))---)............(((......)))..)))).)))))....)).)))).....(((((((((((((......))))))))))))).. ( -38.50, z-score = -3.31, R) >droSec1.super_21 57058 117 - 1102487 GACUUUGUGGUCAACGGGUGGCGAUUCUCC---CAAAAGUACGAUAUCCUAUACUGGAAAACCCUUUGAUAUUAAGCAGUCCAUUCGGCCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAU ((((((...(((((.((((((.(.....))---)............(((......)))..)))).)))))....)).)))).....(((((((((((((......))))))))))))).. ( -37.90, z-score = -2.86, R) >droEre2.scaffold_4690 4194805 117 + 18748788 GACCUUGUGGCCAAAGGCAGGCAAAUCCAC---CGGUAGUACAAUAUCCUAUACCGGAAGUGCCUACGAUAUUAAGAGGUCCAUUCGACCCUUUGAUUUGAUCAAAAGUGGCAGGGUCAU ((((((((.((...(((((((.....)).(---(((((.............))))))...))))).(((.((((((.((((.....)))).))))))))).......)).)))))))).. ( -40.42, z-score = -3.21, R) >droYak2.chrX 6575953 120 - 21770863 GACUUUGUGGCCAAAGGCUAGCAAUUCGCCUGGUGGUAGUACGAUAUCCUAUACCGGAAGUCCCUACGAUAUUAAGAAGUCCACUCGACCCCAUGAUUUGAUCAAAAGUGGUAGGGUCAU ((((((...((((.((((.........))))..)))).(((.(((.(((......))).)))..)))........)))))).....(((((.((.((((......)))).)).))))).. ( -34.70, z-score = -1.29, R) >droAna3.scaffold_13335 2282248 117 + 3335858 CAGCAUGUGGUCCUUGGAGGGCGACAGGCC---CGAGUGCAUCAUCUCCUACACGGGACAGGGCUACGAUUUGCGGAACACCCGACGCCCUCUGAAGCUUCUGAGGAGCGGGCGUGCCAU ..((((...(((((..(((((((...((((---(((.....))...((((....))))..)))))........(((.....))).)))))))..).((((.....))))))))))))... ( -46.30, z-score = -0.73, R) >consensus GACCUUGUGGUCAACGGGUGGCGAUUCCCC___CAAAAGUACGAUAUCCUAUACCGGAAAACCCUACGAUAUUAAGAAGUCCAUUCGACCCUAUCAUUUGAUCAAAAGUGGUAGGGUCAU ((((..((.((((.....)))).)).....................(((......)))...................)))).....((((((((.((((......)))).)))))))).. (-16.84 = -17.90 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:11 2011