| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,260,836 – 12,260,934 |
| Length | 98 |
| Max. P | 0.878667 |

| Location | 12,260,836 – 12,260,934 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.31053 |
| G+C content | 0.41990 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
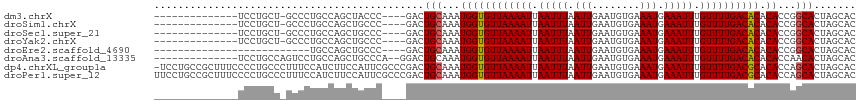
>dm3.chrX 12260836 98 - 22422827 --------------UCCUGCU-GCCCUGCCAGCUACCC----GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC --------------...((((-(...((((.((.....----....))...((((((((((((.(((((.(((........))).))))).)))))))))).))..)))).))))). ( -24.70, z-score = -2.77, R) >droSim1.chrX 9423780 98 - 17042790 --------------UCCUGCU-GCCCUGCCAGCUGCCC----GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC --------------...((((-(...((((...(((..----....)))..((((((((((((.(((((.(((........))).))))).)))))))))).))..)))).))))). ( -25.10, z-score = -2.42, R) >droSec1.super_21 21432 98 - 1102487 --------------UCCUGCU-GCCCUGCCAGCUGCCC----GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC --------------...((((-(...((((...(((..----....)))..((((((((((((.(((((.(((........))).))))).)))))))))).))..)))).))))). ( -25.10, z-score = -2.42, R) >droYak2.chrX 6539865 98 - 21770863 --------------UCCUGCU-GCCCUGCCAGCUGCCC----GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC --------------...((((-(...((((...(((..----....)))..((((((((((((.(((((.(((........))).))))).)))))))))).))..)))).))))). ( -25.10, z-score = -2.42, R) >droEre2.scaffold_4690 4160533 87 + 18748788 --------------------------UGCCAGCUGCCC----GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC --------------------------(((.((.((((.----(.(.......)((((((((((.(((((.(((........))).))))).))))))))))...).)))))).))). ( -21.00, z-score = -1.83, R) >droAna3.scaffold_13335 2244510 101 + 3335858 --------------UCCUGCCAGUCCUGCCAGCUGCCCA--GGACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCAACACUAGCAC --------------...(((((((((((.........))--))))))....((((((((((((.(((((.(((........))).))))).)))))))))).)).........))). ( -26.30, z-score = -2.37, R) >dp4.chrXL_group1a 2966621 116 - 9151740 -UCCUGCCGCUUUCCCCUGCCCUUUCCAUCUUCCAUUCGCCCGACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACGCACACCAGCACUAGCAC -.......(((.....(((..............((((.((......)).))))((((((((((.(((((.(((........))).))))).))))))))))....)))....))).. ( -20.70, z-score = -1.24, R) >droPer1.super_12 2115894 117 + 2414086 UUCCUGCCGCUUUCCCCUGCCCUUUCCAUCUUCCAUUCGCCCGACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACGCACACCAGCACUAGCAC ........(((.....(((..............((((.((......)).))))((((((((((.(((((.(((........))).))))).))))))))))....)))....))).. ( -20.70, z-score = -1.25, R) >consensus ______________UCCUGCU_GCCCUGCCAGCUGCCC____GACUGCAAAUGGUGUUAAAAUUAAUUUAAUUGAAUGUGAAAUGAAAUUUGUUUUGACACACACCGGCACUAGCAC .............................................(((...((((((((((((.(((((.(((........))).))))).)))))))))).))...)))....... (-15.71 = -15.65 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:03 2011