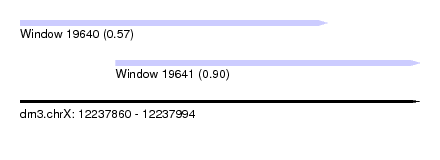
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,237,860 – 12,237,994 |
| Length | 134 |
| Max. P | 0.897362 |

| Location | 12,237,860 – 12,237,963 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Shannon entropy | 0.34367 |
| G+C content | 0.47039 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.07 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
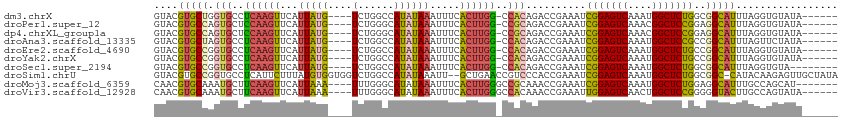
>dm3.chrX 12237860 103 + 22422827 GUACGUGCUGGUGCCUCAAGUUCAUUAUG----UCUGGCCAUAUAAAUUUCACUUGG-CCACAGACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCAUUUAGGUGUAUA------ (((((....((((((.............(----((((((((.............)))-))..)))).....((((((((....)))))))).))))))....))))).------ ( -31.72, z-score = -1.59, R) >droPer1.super_12 2085719 103 - 2414086 GUACGUGCCAGUGCUCCAAGUUCAUUAUG----UCUGGGCAUAUAAAUUUCACUUGG-CCGCAGACCGAAAUCGGAGUCAAACGGCUCCGGAGGCAUUUAGGUGUAUA------ ((((..(((((((((((...........(----(((((((................)-)).))))).......((((((....))))))))).)))))..))))))).------ ( -33.09, z-score = -1.92, R) >dp4.chrXL_group1a 2937101 103 + 9151740 GUACGUGCCAGUGCUCCAAGUUCAUUAUG----UCUGGGCAUAUAAAUUUCACUUGG-CCGCAGACCGAAAUCGGAGUCAAACGGCUCCGGAGGCAUUUAGGUGUAUA------ ((((..(((((((((((...........(----(((((((................)-)).))))).......((((((....))))))))).)))))..))))))).------ ( -33.09, z-score = -1.92, R) >droAna3.scaffold_13335 2216739 103 - 3335858 GUACGUGCUAGUGCCUCAAGUUCAUUAUG----UCUGGCCAUAUAAAUUUCACUUGG-CCACAGACCGAAAUCGGAGUCAAAUGGCUCCGCCGGCAUUUAGUUCUAUA------ ....(.(((((((((.............(----((((((((.............)))-))..))))......(((((((....)))))))..)))).))))).)....------ ( -31.72, z-score = -2.88, R) >droEre2.scaffold_4690 4139092 103 - 18748788 GUACGUGCCGGUGCCUCAAGUUCAUUAUG----UCUGGCCAUAUAAAUUUCACUUGG-CCACAGACCGAAAUCGGAGUCAAAUGGCUCUGCCGGCAUUUAGGUGUAUA------ ((((((((((((................(----((((((((.............)))-))..)))).......((((((....))))))))))))))......)))).------ ( -32.32, z-score = -1.83, R) >droYak2.chrX 20144148 103 - 21770863 GUACGUGCCGGUGCCUCAAGUUCAUUAUG----UCUGGCCAUAUAAAUUUCACUUGG-CCACAGACCGAAAUCGGAGUCAAAUGGCUCUGCCGGCAUUUAGGUGUAUA------ ((((((((((((................(----((((((((.............)))-))..)))).......((((((....))))))))))))))......)))).------ ( -32.32, z-score = -1.83, R) >droSec1.super_2194 4377 101 - 4949 GUACGUGCCGGUGCCUCAAGUUCAUUAUG----UCUGGCCAUAUAAAUUUCACUUGG-CCACAGACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCAUUUAGGUGUA-------- ....(..((.(((((.............(----((((((((.............)))-))..)))).....((((((((....)))))))).)))))...))..).-------- ( -32.72, z-score = -1.78, R) >droSim1.chrU 2400205 111 - 15797150 GUACGUGCCGGUGCCUCAUUCUUUAUGUGGUGGUCUGGCCAUAUAAAUU--GCUGAACCGUCCCACCGAAAUCGGAGUCAAAUGGCUCUGGCGGC-CAUACAAGAGUUGCUAUA (((.(.(((((((((.(((.....))).)).(((..(((.((....)).--)))..)))....))))....((((((((....)))))))).)))-).)))............. ( -33.00, z-score = -0.48, R) >droMoj3.scaffold_6359 413791 103 + 4525533 CAACGUGCAAAUGCUUCAAGUUCAUUAAA----UUUGGGCAUAUAAAUUUCACUUGGGCCGCAAACCGAAAUCGGAGUCAAAUGGCUCUGGAGGCAUUUGCCAGCAU------- ....(.(((((((((((..(((((.....----..))))).............((((........))))...(((((((....))))))))))))))))))).....------- ( -33.70, z-score = -3.11, R) >droVir3.scaffold_12928 3838579 104 + 7717345 CAACGUGCAAAUGCUUCAAGUUCAUUAAA----UUUGGGCAUAUAAAUUUCACUUGGGCCACAAACCGAAAUUGGAGUCAACUGGCUCCGGGGGUACUUGCCAGUAUA------ ....(.((((.((((((..(((((.....----..))))).....((((((..(((.....)))...))))))((((((....)))))).)))))).)))))......------ ( -27.40, z-score = -1.12, R) >consensus GUACGUGCCAGUGCCUCAAGUUCAUUAUG____UCUGGCCAUAUAAAUUUCACUUGG_CCACAGACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCAUUUAGGUGUAUA______ ....(((((.(((..((((((...(((((............))))).....))))))..)))..........(((((((....)))))))..)))))................. (-18.23 = -18.07 + -0.16)
| Location | 12,237,892 – 12,237,994 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Shannon entropy | 0.24044 |
| G+C content | 0.47815 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -23.15 |
| Energy contribution | -24.17 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
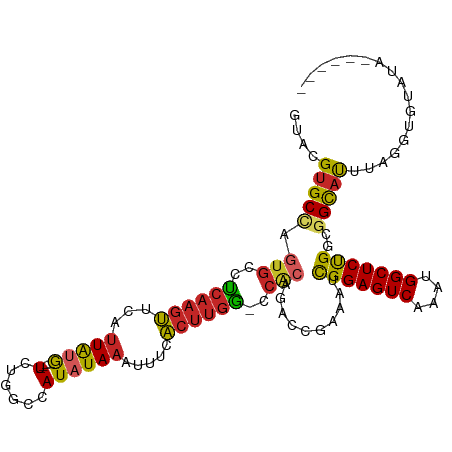
>dm3.chrX 12237892 102 + 22422827 GGCCAUAUAAAUUUCACUUGGCCA-CAGACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCAUUU----AGGU-GUAUAAUAUGCCACGUGCACUG--GCACUAACACACUA--- (((((.............))))).-..(.(((...((((((((....))))))))))))....----.(((-((......(((((.(....)))--))).....))))).--- ( -31.42, z-score = -2.27, R) >droAna3.scaffold_13335 2216771 104 - 3335858 GGCCAUAUAAAUUUCACUUGGCCA-CAGACCGAAAUCGGAGUCAAAUGGCUCCGCCGGCAUUU----AGUU-CUAUAAUAUGCCACGUGCACACACACACACACACACUA--- (((((.............))))).-...........(((((((....)))))))..(((((((----(...-...))).)))))..(((......)))............--- ( -27.92, z-score = -3.41, R) >droEre2.scaffold_4690 4139124 102 - 18748788 GGCCAUAUAAAUUUCACUUGGCCA-CAGACCGAAAUCGGAGUCAAAUGGCUCUGCCGGCAUUU----AGGU-GUAUAAUAUGCCACGUGCAUCG--GCACUAACACACUA--- (((((.............))))).-..(.((((...(((((((....)))))))...((((..----.(((-(((...))))))..)))).)))--))............--- ( -30.02, z-score = -2.03, R) >droYak2.chrX 20144180 107 - 21770863 GGCCAUAUAAAUUUCACUUGGCCA-CAGACCGAAAUCGGAGUCAAAUGGCUCUGCCGGCAUUU----AGGU-GUAUAAUAUGCCACGUGCACUGCCACACCAACACACUGACA (((((.............))))).-(((.(((....(((((((....))))))).))).....----.(((-((......(((.....))).....)))))......)))... ( -30.32, z-score = -1.65, R) >droSec1.super_2194 4409 100 - 4949 GGCCAUAUAAAUUUCACUUGGCCA-CAGACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCAUUU----AGGU---GUAAUAUGCCACGUGCACUG--GCACUAACACACUA--- (((((.............))))).-..(.(((...((((((((....))))))))))))...(----((((---((....(((((.(....)))--)))...)))).)))--- ( -31.82, z-score = -2.47, R) >droSim1.chrU 2400241 106 - 15797150 GGCCAUAUAAAUU--GCUGAACCGUCCCACCGAAAUCGGAGUCAAAUGGCUCUGGCGGCCAUACAAGAGUUGCUAUAAUAUGCCACGUGCACUG--GCACUAACACACUA--- ((((.......((--(.((........)).)))..((((((((....)))))))).))))........((.((........)).))((((....--))))..........--- ( -27.10, z-score = -0.50, R) >consensus GGCCAUAUAAAUUUCACUUGGCCA_CAGACCGAAAUCGGAGUCAAAUGGCUCUGCCGGCAUUU____AGGU_GUAUAAUAUGCCACGUGCACUG__GCACUAACACACUA___ (((((.............))))).............(((((((....)))))))..(((((..................)))))..((((......))))............. (-23.15 = -24.17 + 1.03)

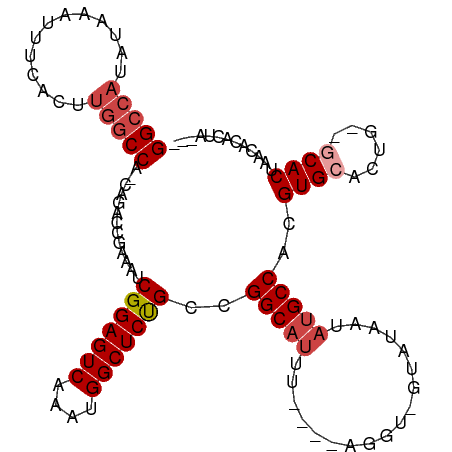
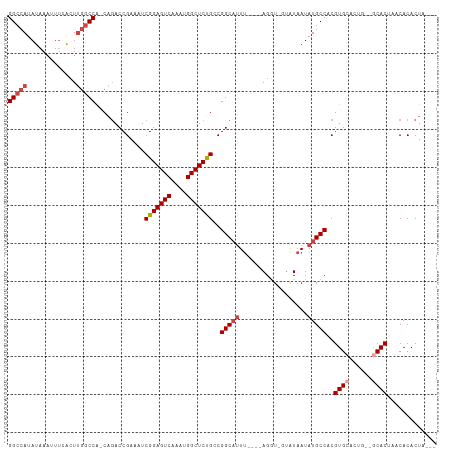
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:59 2011