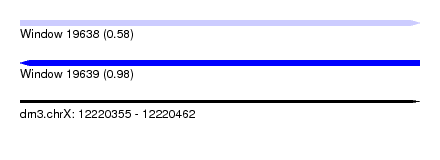
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,220,355 – 12,220,462 |
| Length | 107 |
| Max. P | 0.980806 |

| Location | 12,220,355 – 12,220,462 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Shannon entropy | 0.41423 |
| G+C content | 0.46436 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
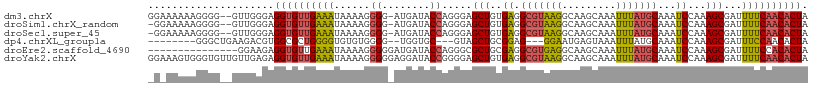
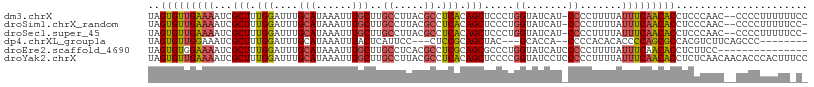
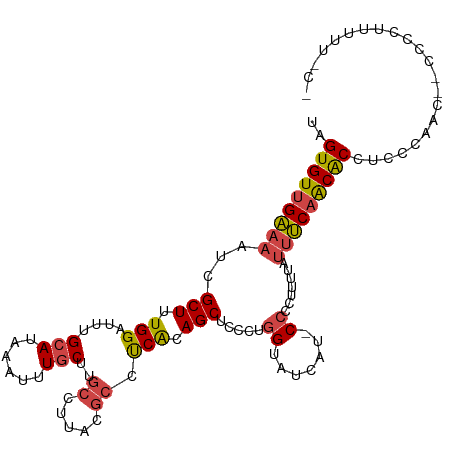
>dm3.chrX 12220355 107 + 22422827 GGAAAAAAGGGG--GUUGGGAGGUGUUGAAAUAAAAGGGG-AUGAUACCAGGGAGCUGUGAGGCGUAAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCAACACUA ............--.......((((((((((.......((-((....((((....))).)..(((((((.........)))))))..))))........)))))))))). ( -24.46, z-score = -1.53, R) >droSim1.chrX_random 3365133 106 + 5698898 -GGAAAAAGGGG--GUUGGGAGGUGUUGAAAUAAAAGGGG-AUGAUACCAGGGAGCUGUGAGGCGUAAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCAACACUA -...........--.......((((((((((.......((-((....((((....))).)..(((((((.........)))))))..))))........)))))))))). ( -24.46, z-score = -1.60, R) >droSec1.super_45 235631 106 + 245362 -GGAAAAAGGGG--GUUGGGAGGUGUUGAAAUAAAAGGGG-AUGAUACCAGGGAGCUGUGAGGCGUAAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCAACACUA -...........--.......((((((((((.......((-((....((((....))).)..(((((((.........)))))))..))))........)))))))))). ( -24.46, z-score = -1.60, R) >dp4.chrXL_group1a 2911407 94 + 9151740 --------GGGCUGAAGACGUGGCGCUGGGGUGUGUGGGG--UGGUGC---GUAGCUGCGGAG---GGAAUGAGUAAAUUUAUGCAAAUCCAAAGCGAUUUCCAACACUA --------(.(((........))).)(((((..(((.(((--(..(((---((((.(((....---.......)))...))))))).))))...)))..)))))...... ( -21.30, z-score = 1.73, R) >droEre2.scaffold_4690 4122969 95 - 18748788 ---------------GGAAGAGGUGUUGAAAUAAAAGGGGGAUGAUACCAGGGCGCUGCGAGGCGUGAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCCACACUA ---------------((((((((((((................))))))....((((..((.(((((((.........)))))))...))...)))).))))))...... ( -23.09, z-score = -0.35, R) >droYak2.chrX 20128991 110 - 21770863 GGAAAGUGGGUGUUGUUGAGAGGUGUUGAAAUAAAAGGGGGAGGAUACCGGGGAGCUGUGAGGCGUAAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCAACACUA .....................((((((((((......((........)).....(((..((.(((((((.........)))))))...))...)))...)))))))))). ( -24.00, z-score = -0.59, R) >consensus _G_AAAAAGGGG__GUUGGGAGGUGUUGAAAUAAAAGGGG_AUGAUACCAGGGAGCUGUGAGGCGUAAGGCAAGCAAAUUUAUGCAAAUCCAAAGCGAUUUUCAACACUA .....................((((((((((......((........)).....(((..((.(((((((.........)))))))...))...)))...)))))))))). (-19.59 = -19.57 + -0.02)
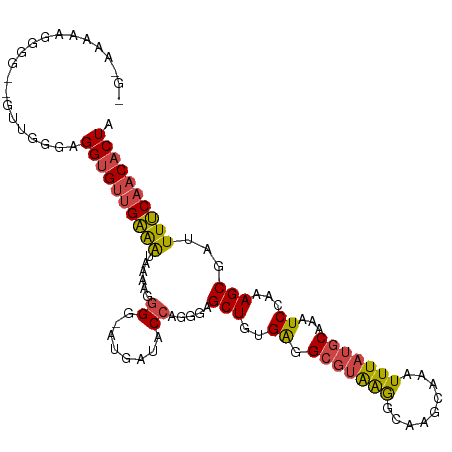
| Location | 12,220,355 – 12,220,462 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Shannon entropy | 0.41423 |
| G+C content | 0.46436 |
| Mean single sequence MFE | -17.74 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12220355 107 - 22422827 UAGUGUUGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUUACGCCUCACAGCUCCCUGGUAUCAU-CCCCUUUUAUUUCAACACCUCCCAAC--CCCCUUUUUUCC ..(((((((((........((((..(((......))).((((....((......)).....))))..))-)).......)))))))))........--............ ( -18.96, z-score = -2.90, R) >droSim1.chrX_random 3365133 106 - 5698898 UAGUGUUGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUUACGCCUCACAGCUCCCUGGUAUCAU-CCCCUUUUAUUUCAACACCUCCCAAC--CCCCUUUUUCC- ..(((((((((........((((..(((......))).((((....((......)).....))))..))-)).......)))))))))........--...........- ( -18.96, z-score = -2.99, R) >droSec1.super_45 235631 106 - 245362 UAGUGUUGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUUACGCCUCACAGCUCCCUGGUAUCAU-CCCCUUUUAUUUCAACACCUCCCAAC--CCCCUUUUUCC- ..(((((((((........((((..(((......))).((((....((......)).....))))..))-)).......)))))))))........--...........- ( -18.96, z-score = -2.99, R) >dp4.chrXL_group1a 2911407 94 - 9151740 UAGUGUUGGAAAUCGCUUUGGAUUUGCAUAAAUUUACUCAUUCC---CUCCGCAGCUAC---GCACCA--CCCCACACACCCCAGCGCCACGUCUUCAGCCC-------- ..((((((((((((......)))))...................---....((......---))....--...........)))))))..............-------- ( -13.80, z-score = 0.19, R) >droEre2.scaffold_4690 4122969 95 + 18748788 UAGUGUGGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUCACGCCUCGCAGCGCCCUGGUAUCAUCCCCCUUUUAUUUCAACACCUCUUCC--------------- ..((((.((((........((((..(((......))).((((...(((......)))....))))..))))........)))).)))).......--------------- ( -17.89, z-score = -0.58, R) >droYak2.chrX 20128991 110 + 21770863 UAGUGUUGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUUACGCCUCACAGCUCCCCGGUAUCCUCCCCCUUUUAUUUCAACACCUCUCAACAACACCCACUUUCC ..(((((((((........(((...(((......))).((((....((......)).....))))...)))........)))))))))...................... ( -17.89, z-score = -2.50, R) >consensus UAGUGUUGAAAAUCGCUUUGGAUUUGCAUAAAUUUGCUUGCCUUACGCCUCACAGCUCCCUGGUAUCAU_CCCCUUUUAUUUCAACACCUCCCAAC__CCCCUUUUU_C_ ..(((((((((...(((.(((....(((......)))..((.....)).))).))).....((........))......)))))))))...................... (-13.31 = -13.57 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:57 2011