
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,202,618 – 12,202,717 |
| Length | 99 |
| Max. P | 0.785008 |

| Location | 12,202,618 – 12,202,717 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Shannon entropy | 0.53433 |
| G+C content | 0.53046 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
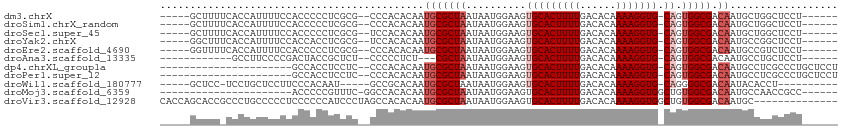
>dm3.chrX 12202618 99 + 22422827 -----GCUUUUCACCAUUUUCCACCCCCUCGCG--CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCUGGCUCCU------ -----((((.((.((((((((((.......(((--(.........))))......))))))(((((((((......)))))))-)))))).)).)).))........------ ( -27.02, z-score = -1.31, R) >droSim1.chrX_random 3339816 99 + 5698898 -----GCUUUUCACCAUUUUCCACCCCCUCGCG--CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCUGGCUCCU------ -----((((.((.((((((((((.......(((--(.........))))......))))))(((((((((......)))))))-)))))).)).)).))........------ ( -27.02, z-score = -1.31, R) >droSec1.super_45 217965 99 + 245362 -----GCUUUUCACCAUUUUCCACCCCCUCGCG--UCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCUGGCUCCU------ -----((((.((.((((((((((......((((--(.......))))).......))))))(((((((((......)))))))-)))))).)).)).))........------ ( -26.32, z-score = -1.17, R) >droYak2.chrX 20111603 99 - 21770863 -----GGCUUUCACCAUUUUCCACCACCUCGCG--UCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCCGGCUCCU------ -----(((..((.((((((((((......((((--(.......))))).......))))))(((((((((......)))))))-)))))).))....))).......------ ( -29.62, z-score = -1.86, R) >droEre2.scaffold_4690 4106087 99 - 18748788 -----GGUUUUCACCAUUUUCCACCCCCUCGCG--CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCCGUCUCCU------ -----(((..((.((((((((((.......(((--(.........))))......))))))(((((((((......)))))))-)))))).))....))).......------ ( -28.72, z-score = -2.11, R) >droAna3.scaffold_13335 2183295 89 - 3335858 ------------GCCUUCCCCGACUACCGCUCU--CCCCCUCU---CGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCCUGCUCCU------ ------------(((((((.........((...--........---.)).......)))))(((((((((......)))))))-))..(((......))).))....------ ( -18.49, z-score = -0.28, R) >dp4.chrXL_group1a 2890017 88 + 9151740 ----------------------GCCACCUCCUC--CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCCUCGCCCUGCUCCU ----------------------...........--......(((((((((.........))))))...))).......(((.(-(((.(((((.......))))))))).))) ( -24.40, z-score = -1.86, R) >droPer1.super_12 2040448 88 - 2414086 ----------------------GCCACCUCCUC--CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGUGGCGACAAUGCCUCGCCCUGCUCCU ----------------------...........--......(((((((((.........))))))...))).......(((.(-(((.(((((.......))))))))).))) ( -24.40, z-score = -1.86, R) >droWil1.scaffold_180777 3959445 91 + 4753960 -----GCUCC-UCCUGCUCCUUCCCACAAU-----GCCGCACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG-CAGGGGCGACAAUACACCU---------- -----(((((-........(((((......-----((.((.....)))).......)))))(((((((((......)))))))-)))))))............---------- ( -28.22, z-score = -2.44, R) >droMoj3.scaffold_6359 359979 84 + 4525533 ----------------------ACCCCCGUUUC-GGCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUGGCUGUGGCGACAAUGCCAACCGCC------ ----------------------.....(((..(-((((((....((((((.........))))))((((((...)))))))))))))..)))...............------ ( -26.70, z-score = -1.36, R) >droVir3.scaffold_12928 3784449 99 + 7717345 CACCAGCACCGCCCUGCCCCCUCCCCCCAUCCCUAGCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUGGCUGUGGCGACAAUGC-------------- .....(((.(((((.(((.(((....((((...((((((.....)).))))...))))..(((((....)).)))...))).))).).))))....)))-------------- ( -26.70, z-score = -1.10, R) >consensus _____GCUUU_CACCAUUUUCCACCCCCUCGCG__CCCACACAAUGCGCUAAUAAUGGAAGUGCACUUUUGACACAAAAGGUG_CAGUGGCGACAAUGCCGGCUCCU______ ....................................((((....((((((.........))))))((((((...))))))......))))....................... (-12.64 = -13.00 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:54 2011