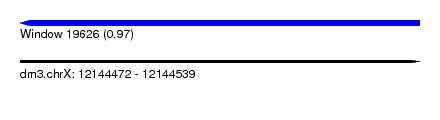
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,144,472 – 12,144,539 |
| Length | 67 |
| Max. P | 0.966910 |

| Location | 12,144,472 – 12,144,539 |
|---|---|
| Length | 67 |
| Sequences | 12 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 90.39 |
| Shannon entropy | 0.22275 |
| G+C content | 0.46915 |
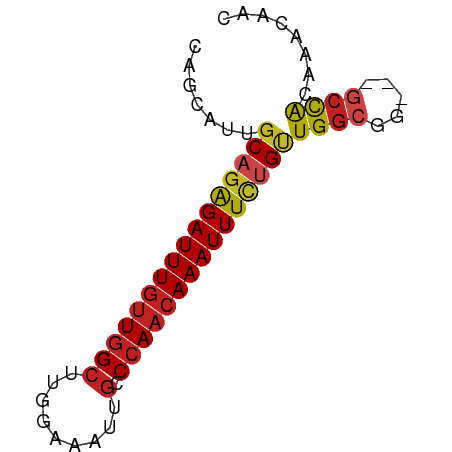
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
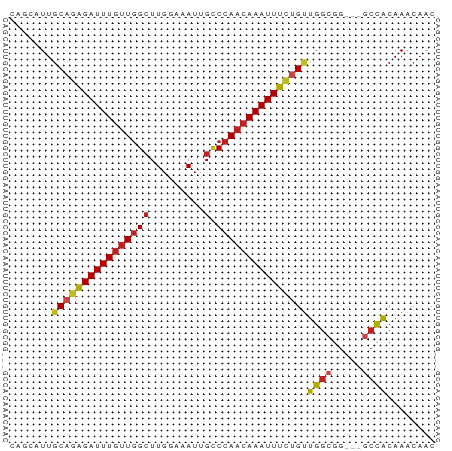
>dm3.chrX 12144472 67 - 22422827 CAGCAUUGCAGAGAUUUGCUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---GCCACAAACAAC ..((...(((((((((((..(((.........)))...)))))))))))..))..---............ ( -20.10, z-score = -1.35, R) >droSim1.chrX_random 3311712 67 - 5698898 CAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---GCCACAAACAAC ..((...((((((((((((((((.........).)))))))))))))))..))..---............ ( -23.80, z-score = -2.83, R) >droSec1.super_45 162320 67 - 245362 CAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---GCCACAAACAAC ..((...((((((((((((((((.........).)))))))))))))))..))..---............ ( -23.80, z-score = -2.83, R) >droYak2.chrX 20054797 67 + 21770863 CAGCAUUGCAGAGAUUUGUUGGGUUGGAAAUUGGCCAACAAAUUUCGGUUGGCGG---GCCACAAACAAC ..((...((.((((((((((((.(........).)))))))))))).))..))..---............ ( -20.20, z-score = -2.01, R) >droEre2.scaffold_4690 4050662 67 + 18748788 CGGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCGGUUGGCGG---GCUACAAACAAC .(((.((((.(((((((((((((.........).)))))))))))).....))))---)))......... ( -20.80, z-score = -2.15, R) >droAna3.scaffold_13335 2121510 70 + 3335858 CGACAUUGCAGGGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGGGCUGCCGCAAGCAGC ((.(...((((..((((((((((.........).)))))))))..))))..))).((((.(....))))) ( -27.80, z-score = -2.38, R) >dp4.Unknown_singleton_1895 28379 67 + 56477 CAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---CCCACAAACAAC ..((...((((((((((((((((.........).)))))))))))))))..))..---............ ( -23.80, z-score = -2.91, R) >droPer1.super_12 1978435 67 + 2414086 CACCAUUCCAGAGAUUUGUUGGCUUGGAAAUUGCCCAAAAAAUUUUUGUUGGCGG---GCCACAAGCAAC .....................(((((....(((((((((.....))))..)))))---....)))))... ( -13.80, z-score = 0.67, R) >droWil1.scaffold_180777 3902216 64 - 4753960 ---UGUUGCACAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---GCCACAAACUAC ---........((.(((((.((((((........((((((......)))))))))---)))))))))).. ( -17.10, z-score = -0.86, R) >droVir3.scaffold_12928 3709417 67 - 7717345 CAGCUUUGCAGAGAUUUGUUAGCUUGGAAAUUGUCCAACAAAUUUCUGUUGCCGG---CCCACAAACAAC ..(((..((((((((((((.....((((.....))))))))))))))))....))---)........... ( -18.50, z-score = -2.10, R) >droMoj3.scaffold_6359 276993 67 - 4525533 CAGCUUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG---GCCACAAACAAC ..(((..((((((((((((((((.........).))))))))))))))).)))..---............ ( -24.40, z-score = -2.97, R) >droGri2.scaffold_15081 249730 62 + 4274704 -----UUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGCUGGCGG---GCCACAAACAGC -----..((((((((((((((((.........).)))))))))))))))(((...---.)))........ ( -24.10, z-score = -3.02, R) >consensus CAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGCGG___GCCACAAACAAC .......((((((((((((((((.........).)))))))))))))))((((.....))))........ (-19.08 = -19.63 + 0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:47 2011