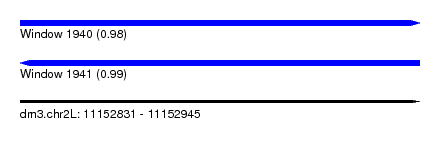
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,152,831 – 11,152,945 |
| Length | 114 |
| Max. P | 0.990053 |

| Location | 11,152,831 – 11,152,945 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 65.15 |
| Shannon entropy | 0.74883 |
| G+C content | 0.48056 |
| Mean single sequence MFE | -37.91 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
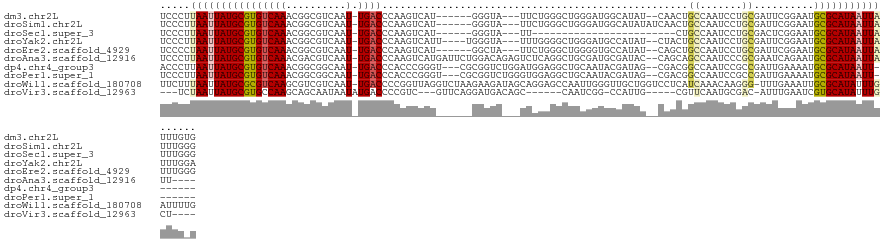
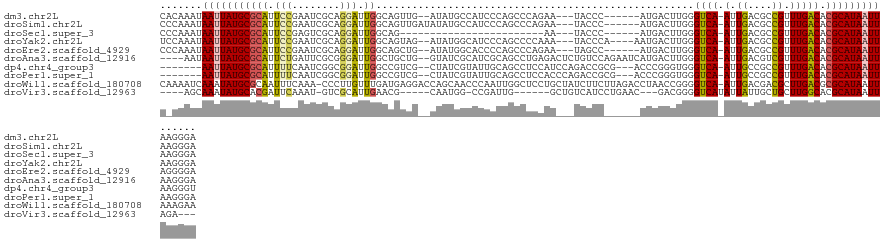
>dm3.chr2L 11152831 114 + 23011544 UCCCUUAAUUAUGCGUGUCAAACGGCGUCAAU-UGACCCAAGUCAU------GGGUA---UUCUGGGCUGGGAUGGCAUAU--CAACUGCCAAUCCUGCGAUUCGGAAUGCGCAUAAUUAUUUGUG .....((((((((((((((....)))......-..(((((.....)------))))(---((((((((.(((((((((...--....)))).)))))))...))))))))))))))))))...... ( -38.00, z-score = -2.30, R) >droSim1.chr2L 10952766 116 + 22036055 UCCCUUAAUUAUGCGUGUCAAACGGCGUCAAU-UGACCCAAGUCAU------GGGUA---UUCUGGGCUGGGAUGGCAUAUAUCAACUGCCAAUCCUGCGAUUCGGAAUGCGCAUAAUUAUUUGGG .(((.((((((((((((((....)))......-..(((((.....)------))))(---((((((((.(((((((((.........)))).)))))))...))))))))))))))))))...))) ( -41.90, z-score = -3.05, R) >droSec1.super_3 6554110 92 + 7220098 UCCCUUAAUUAUGCGUGUCAAACGGCGUCAAU-UGACCCAAGUCAU------GGGUA---UU------------------------CUGCCAAUCCUGCGACUCGGAAUGCGCAUAAUUAUUUGGG .(((.(((((((((((((.....((((.....-..(((((.....)------)))).---..------------------------.))))..(((........))))))))))))))))...))) ( -31.50, z-score = -3.35, R) >droYak2.chr2L 7549348 116 + 22324452 UCCCUUAAUUAUGCGUGUCAAACGGCGUCAAU-UGACCCAAGUCAUU----UGGGUA---UUUGGGGCUGGGAUGCCAUAU--CUACUGCCAAUCCUGCGAUUCGGAAUGCGCAUAAUUAUUUGGA (((..((((((((((((((...((((.((((.-..(((((((...))----))))).---.)))).)))).))))).....--.....((...(((........)))..)))))))))))...))) ( -39.30, z-score = -2.20, R) >droEre2.scaffold_4929 12350803 114 - 26641161 UCCCCUAAUUAUGCGUGUCAAACGGCGUCAAU-UGACCCAAGUCAU------GGCUA---UUCUGGGCUGGGGUGCCAUAU--CAGCUGCCAAUCCUGCGAUUCGGAAUGCGCAUAAUUAUUUGGG .(((.(((((((((((((.....(((((((..-((((....)))))------)))..---.....((((((.........)--))))))))..(((........))))))))))))))))...))) ( -36.80, z-score = -1.05, R) >droAna3.scaffold_12916 11883317 119 - 16180835 UCCCUUAAUUAUGCGUGUCAAACGACGUCAAU-UGACCCAAGUCAUGAUUCUGGACAGAGUCUCAGGCUGCGAUGCGAUAC--CAGCAGCCAAUCCCGCGAAUCAGAAUGCGCAUAAUUAUU---- .....((((((((((((((....((((((...-.)))....))).(((((((((...(.....).((((((..........--..))))))....))).))))))).)))))))))))))..---- ( -37.50, z-score = -3.40, R) >dp4.chr4_group3 6514266 113 + 11692001 ACCCUUAAUUAUGCGUGUCAAACGGCGGCAAU-UGACCCACCCGGGU---CGCGGUCUGGAUGGAGGCUGCAAUACGAUAG--CGACGGCCAAUCCGCCGAUUGAAAAUGCGCAUAAUU------- ......((((((((((.((((.((((((....-.(((((....))))---)(((((((......)))))))..........--...........)))))).))))....))))))))))------- ( -45.90, z-score = -3.01, R) >droPer1.super_1 8005689 113 + 10282868 UCCCUUAAUUAUGCGUGUCAAACGGCGGCAAU-UGACCCACCCGGGU---CGCGGUCUGGGUGGAGGCUGCAAUACGAUAG--CGACGGCCAAUCCGCCGAUUGAAAAUGCGCAUAAUU------- ......((((((((((.((((.((((((....-....(((((((((.---.....))))))))).((((((..........--.).)))))...)))))).))))....))))))))))------- ( -49.00, z-score = -3.37, R) >droWil1.scaffold_180708 7923920 124 - 12563649 UUCUUUAAUUAUGCGCGUCAAGCGUCGUCAAU-UGACCCCGGUUAGGUCUAAGAAGAUAGCAGGAGCCAAUUGGGUUGCUGGUCCUCAUCAAACAAGGG-UUUGAAAUUGCGCAUAUUUGAUUUUG ....((((.((((((((((((((((((.....-))))((..(((.(((....((.(((..(((.((((.....)))).)))))).))))).)))..)))-)))))...)))))))).))))..... ( -36.10, z-score = -1.64, R) >droVir3.scaffold_12963 17861982 103 - 20206255 ---UCUAAUUAUGCGUGCCAAGCAGCAAUAAUAUGACCCCGUC---GUUCAGGAUGACAGC------CAAUCGG-CCAUUG-----CGUUCAAUGCGAC-AUUUGAAUCGUGCAUAUUUGCU---- ---..(((.(((((((((......))).............(((---((...(((((.((((------(....))-)...))-----)))))...)))))-..........)))))).)))..---- ( -23.10, z-score = 0.57, R) >consensus UCCCUUAAUUAUGCGUGUCAAACGGCGUCAAU_UGACCCAAGUCAU______GGGUA___UUCGAGGCUGGGAUGCCAUAG__CAACUGCCAAUCCUGCGAUUCGGAAUGCGCAUAAUUAUUUG_G .....((((((((((((((....)))..............................................................((.......))..........)))))))))))...... (-11.25 = -11.39 + 0.14)
| Location | 11,152,831 – 11,152,945 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 65.15 |
| Shannon entropy | 0.74883 |
| G+C content | 0.48056 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.16 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
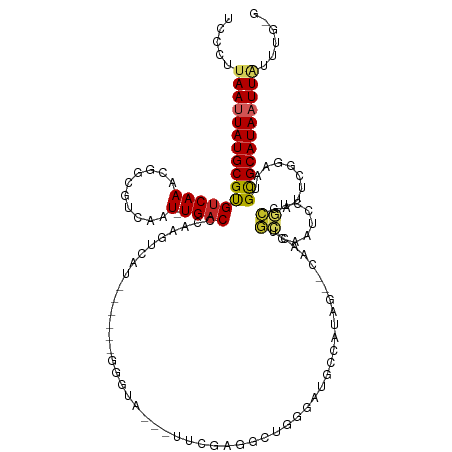
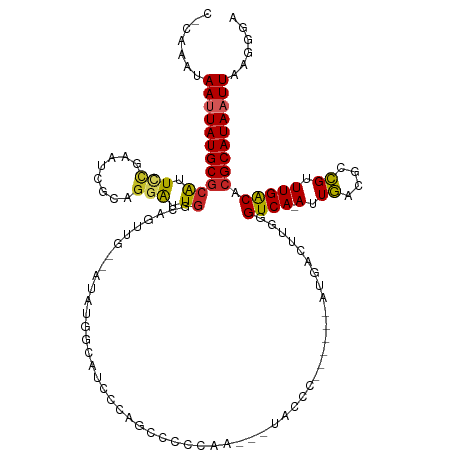
>dm3.chr2L 11152831 114 - 23011544 CACAAAUAAUUAUGCGCAUUCCGAAUCGCAGGAUUGGCAGUUG--AUAUGCCAUCCCAGCCCAGAA---UACCC------AUGACUUGGGUCA-AUUGACGCCGUUUGACACGCAUAAUUAAGGGA ......((((((((((.....(((((.((.((..(((((....--...)))))..))....(((..---.((((------(.....)))))..-.)))..)).)))))...))))))))))..... ( -34.20, z-score = -2.23, R) >droSim1.chr2L 10952766 116 - 22036055 CCCAAAUAAUUAUGCGCAUUCCGAAUCGCAGGAUUGGCAGUUGAUAUAUGCCAUCCCAGCCCAGAA---UACCC------AUGACUUGGGUCA-AUUGACGCCGUUUGACACGCAUAAUUAAGGGA (((...((((((((((.....(((((.((.((..(((((.........)))))..))....(((..---.((((------(.....)))))..-.)))..)).)))))...)))))))))).))). ( -37.40, z-score = -3.04, R) >droSec1.super_3 6554110 92 - 7220098 CCCAAAUAAUUAUGCGCAUUCCGAGUCGCAGGAUUGGCAG------------------------AA---UACCC------AUGACUUGGGUCA-AUUGACGCCGUUUGACACGCAUAAUUAAGGGA (((...((((((((((....((((((((..(((((.....------------------------))---).)).------.))))))))((((-(.((....)).))))).)))))))))).))). ( -32.80, z-score = -3.76, R) >droYak2.chr2L 7549348 116 - 22324452 UCCAAAUAAUUAUGCGCAUUCCGAAUCGCAGGAUUGGCAGUAG--AUAUGGCAUCCCAGCCCCAAA---UACCCA----AAUGACUUGGGUCA-AUUGACGCCGUUUGACACGCAUAAUUAAGGGA (((...((((((((((...(((........)))..(((.....--....(((......))).(((.---.(((((----(.....))))))..-.)))..)))........))))))))))..))) ( -33.50, z-score = -1.74, R) >droEre2.scaffold_4929 12350803 114 + 26641161 CCCAAAUAAUUAUGCGCAUUCCGAAUCGCAGGAUUGGCAGCUG--AUAUGGCACCCCAGCCCAGAA---UAGCC------AUGACUUGGGUCA-AUUGACGCCGUUUGACACGCAUAAUUAGGGGA (((...((((((((((....((((.(((......((((..(((--....(((......))))))..---..)))------)))).))))((((-(.((....)).))))).)))))))))).))). ( -36.60, z-score = -1.73, R) >droAna3.scaffold_12916 11883317 119 + 16180835 ----AAUAAUUAUGCGCAUUCUGAUUCGCGGGAUUGGCUGCUG--GUAUCGCAUCGCAGCCUGAGACUCUGUCCAGAAUCAUGACUUGGGUCA-AUUGACGUCGUUUGACACGCAUAAUUAAGGGA ----..((((((((((((.((((((((...((((.((((((.(--((.....))))))))).........)))).)))))).))..)).((((-(.((....)).))))).))))))))))..... ( -39.60, z-score = -2.06, R) >dp4.chr4_group3 6514266 113 - 11692001 -------AAUUAUGCGCAUUUUCAAUCGGCGGAUUGGCCGUCG--CUAUCGUAUUGCAGCCUCCAUCCAGACCGCG---ACCCGGGUGGGUCA-AUUGCCGCCGUUUGACACGCAUAAUUAAGGGU -------(((((((((.....((((.(((((((((((((((.(--(.........)).))...(((((.(......---..).))))))))))-))..)))))).))))..)))))))))...... ( -40.30, z-score = -1.29, R) >droPer1.super_1 8005689 113 - 10282868 -------AAUUAUGCGCAUUUUCAAUCGGCGGAUUGGCCGUCG--CUAUCGUAUUGCAGCCUCCACCCAGACCGCG---ACCCGGGUGGGUCA-AUUGCCGCCGUUUGACACGCAUAAUUAAGGGA -------(((((((((.....((((.(((((((((((((((.(--(.........)).))...(((((.(......---..).))))))))))-))..)))))).))))..)))))))))...... ( -42.90, z-score = -1.96, R) >droWil1.scaffold_180708 7923920 124 + 12563649 CAAAAUCAAAUAUGCGCAAUUUCAAA-CCCUUGUUUGAUGAGGACCAGCAACCCAAUUGGCUCCUGCUAUCUUCUUAGACCUAACCGGGGUCA-AUUGACGACGCUUGACGCGCAUAAUUAAAGAA ..........(((((((.........-(((..((((((.(((((..((((..((....))....)))).)))))))))))......)))((((-(.((....)).))))))))))))......... ( -32.70, z-score = -2.61, R) >droVir3.scaffold_12963 17861982 103 + 20206255 ----AGCAAAUAUGCACGAUUCAAAU-GUCGCAUUGAACG-----CAAUGG-CCGAUUG------GCUGUCAUCCUGAAC---GACGGGGUCAUAUUAUUGCUGCUUGGCACGCAUAAUUAGA--- ----......(((((.((((......-))))(((((....-----)))))(-((((..(------(((((.((((((...---..)))))).))).....)))..)))))..)))))......--- ( -27.50, z-score = -0.15, R) >consensus C_CAAAUAAUUAUGCGCAUUCCGAAUCGCAGGAUUGGCAGUUG__AUAUGGCAUCCCAGCCCCCAA___UACCC______AUGACUUGGGUCA_AUUGACGCCGUUUGACACGCAUAAUUAAGGGA .......(((((((((((.(((........))).)).....................................................((((...((....))..)))).)))))))))...... (-13.21 = -13.16 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:39 2011