
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,132,450 – 12,132,589 |
| Length | 139 |
| Max. P | 0.649445 |

| Location | 12,132,450 – 12,132,550 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.51427 |
| G+C content | 0.52544 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
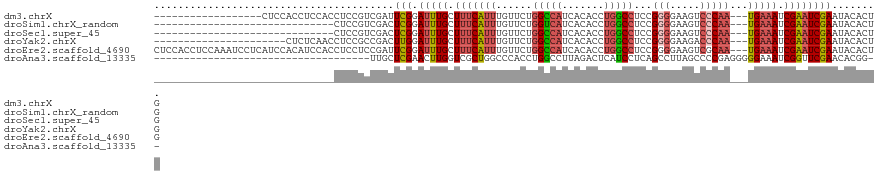
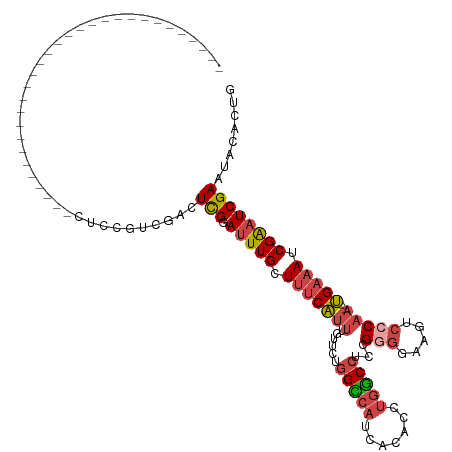
>dm3.chrX 12132450 100 - 22422827 ------------------CUCCACCUCCACCUCCGUCGAUUCGGAUUUGCUUUCAUUUGUUCUGGCCAUCACACCUGGCCUCCGGGGAAGUCCCAA---UGAAAUCGAAUCGAAUACACUG ------------------.................(((((((((......(((((((......(((((.......)))))....(((....)))))---))))))))))))))........ ( -26.70, z-score = -1.72, R) >droSim1.chrX_random 3302805 88 - 5698898 ------------------------------CUCCGUCGACUCGGAUUUGCUUUCAUUUGUUCUGGUCAUCACACCUGGCCUCCGGGGAAGUCCCAA---UGAAAUCGAAUCGAAUACACUG ------------------------------...((......))((((((.(((((((......(((((.......)))))....(((....)))))---))))).)))))).......... ( -21.20, z-score = -0.24, R) >droSec1.super_45 150454 88 - 245362 ------------------------------CUCCGUCGACUCGGAUUUGCUUUCAUUUGUUCUGGCCAUCACACCUGGCCUCCGGGGAAGUCCCAA---UGAAAUCGAAUCGAAUACACUG ------------------------------...((......))((((((.(((((((......(((((.......)))))....(((....)))))---))))).)))))).......... ( -23.90, z-score = -1.25, R) >droYak2.chrX 20042615 96 + 21770863 ----------------------CUCUCAACCUCCGCCGACUUGGAUUUGCUUUCAUUUGUUCUGGCCAUCACACCUGGCCUCCGGGGAAGACCCAA---UGAAAUCGAAUCGAAUACACUG ----------------------..................((.((((((.(((((((......(((((.......)))))...(((.....)))))---))))).)))))).))....... ( -24.80, z-score = -1.84, R) >droEre2.scaffold_4690 4038673 118 + 18748788 CUCCACCUCCAAAUCCUCAUCCACAUCCACCUCCUCCGAUUCGGAUUUGCUUUCAUUUGUUCUGGCCAUCACACCUGGCCUCCGGGGAAGUCGCAA---UGAAAUCGAAUCGAAUACACUG ....................................((((((((..((((....((((.(((.(((((.......)))))...))).)))).))))---.....))))))))......... ( -22.30, z-score = -0.01, R) >droAna3.scaffold_13335 2111863 83 + 3335858 ------------------------------------UUGCUCGAACUUGGUCGCUGGCCCACCUGGCCUUAGACUCAUCCUCAGCCUUAGCCCCGAGGGGGAAAUCGGUUCGAACACGG-- ------------------------------------.((.(((((((..(((...((((.....))))...)))((..((((.((....))...))))..))....))))))).))...-- ( -27.00, z-score = -0.16, R) >consensus ______________________________CUCCGUCGACUCGGAUUUGCUUUCAUUUGUUCUGGCCAUCACACCUGGCCUCCGGGGAAGUCCCAA___UGAAAUCGAAUCGAAUACACUG ........................................(((.(((((.(((((........(((((.......)))))...(((.....))).....))))).))))))))........ (-14.38 = -14.93 + 0.56)
| Location | 12,132,490 – 12,132,589 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.73 |
| Shannon entropy | 0.47541 |
| G+C content | 0.47717 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -13.13 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
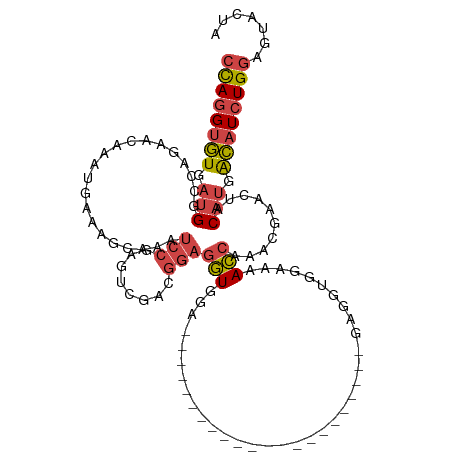
>dm3.chrX 12132490 99 + 22422827 CCAGGUGUGAUGGCCAGAACAAAUGAAAGCAAAUCCGAAUCGACGGAGGUGGA------------------GGUGGAGGUGGAAAACCAAACGAACUACAUGACAUCUGGAGUACUA ((((((((.(((.....................((((..((.((....)).))------------------..))))(((.....)))..........))).))))))))....... ( -28.80, z-score = -2.29, R) >droSim1.chrX_random 3302845 87 + 5698898 CCAGGUGUGAUGACCAGAACAAAUGAAAGCAAAUCCGAGUCGACGGAGGUGGA------------------------------AAACCAAACGAACUACAUGACAUCUGGAGUACUA ((((((((.(((.....................((((......))))(((...------------------------------..)))..........))).))))))))....... ( -24.00, z-score = -2.55, R) >droSec1.super_45 150494 87 + 245362 CCAGGUGUGAUGGCCAGAACAAAUGAAAGCAAAUCCGAGUCGACGGAGGUGGA------------------------------AAACCAAACGAACUACAUGACAUCUGGAGUACUA ((((((((.(((.....................((((......))))(((...------------------------------..)))..........))).))))))))....... ( -24.20, z-score = -2.24, R) >droYak2.chrX 20042655 95 - 21770863 CCAGGUGUGAUGGCCAGAACAAAUGAAAGCAAAUCCAAGUCGGCGGAGGUUGA----------------------GAGGUGGAAAACCAAACGAACUACAUAACAUCUGGAGUACUA ((((((((.(((...............(((...(((........))).)))..----------------------..(((.....)))..........))).))))))))....... ( -21.70, z-score = -0.57, R) >droEre2.scaffold_4690 4038713 117 - 18748788 CCAGGUGUGAUGGCCAGAACAAAUGAAAGCAAAUCCGAAUCGGAGGAGGUGGAUGUGGAUGAGGAUUUGGAGGUGGAGGUGGAAAACCAAACGAACUACAUGACAUCUGGAGUACUA ((((((((.(((.(((((.(........(((..((((..((....))..))))))).......).))))).(((.....(((....))).....))).))).))))))))....... ( -32.16, z-score = -2.17, R) >droAna3.scaffold_13335 2111892 93 - 3335858 CUAAGGCUGAGGAUGAGUCUAAGGCCAGGUGGGCCAG---CGACCAAGUUCGA---------------------GCAAAUGGAAGAGAACAUAAACUACACGGGAUCUGAGGAACUA ....((((..(((....)))..)))).(((.(.....---).))).(((((..---------------------....(((........)))........(((...)))..))))). ( -17.80, z-score = 1.29, R) >consensus CCAGGUGUGAUGGCCAGAACAAAUGAAAGCAAAUCCGAGUCGACGGAGGUGGA______________________GAGGUGGAAAACCAAACGAACUACAUGACAUCUGGAGUACUA ((((((((.(((.....................(((........)))(((...................................)))..........))).))))))))....... (-13.13 = -13.80 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:44 2011