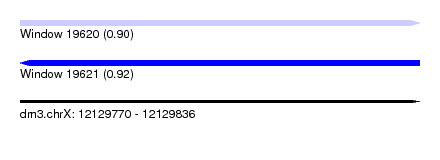
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,129,770 – 12,129,836 |
| Length | 66 |
| Max. P | 0.918894 |

| Location | 12,129,770 – 12,129,836 |
|---|---|
| Length | 66 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.32375 |
| G+C content | 0.45504 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.81 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.897767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
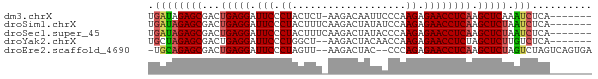
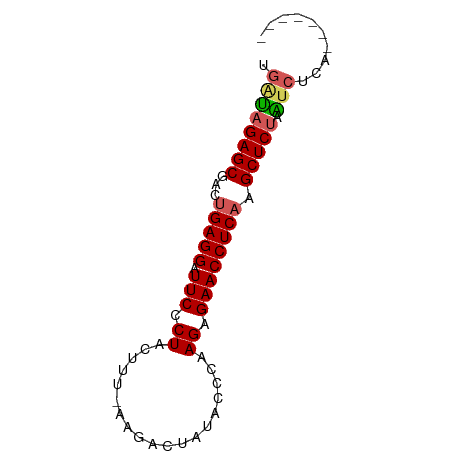
>dm3.chrX 12129770 66 + 22422827 -------UGAGAUUUGAGCUUGAGGUUCUCUUGGGAAUUGUCUU-AGAGUAGGGAAUCCUCAGUCGCUCUAUCA -------...(((..(((((((((((((((((((((....))))-)....)))))).))))))..)))).))). ( -22.00, z-score = -1.64, R) >droSim1.chrX 9351360 67 + 17042790 -------UGAGAUUAGAGCUUGAGGUUCUCUUGGAUAUAGUCUUGAAAGUAGGGAAUCCUCAGUCGCUCUAUCA -------...(((.(((((((((((((((((.((((...)))).......)))))).))))))..)))))))). ( -21.90, z-score = -1.87, R) >droSec1.super_45 147769 67 + 245362 -------UGAGAUUAGAGCUUGAGGUUCUCUUGGGUAUAGUCUUGAAAGUAGGGAAUCCUCAGUCGCUCUAUCA -------...(((.(((((((((((((((((..((......))..).....))))).))))))..)))))))). ( -22.00, z-score = -1.91, R) >droYak2.chrX 20039125 65 - 21770863 -------UGAGACAAGAGCUAGAGGUUCUCUUGGUUGUAGUCUU--AGCCAGGGAAUCCUCAGUCGCUCUAGCA -------.......(((((..(((((((.((((((((......)--)))))))))).))))....))))).... ( -24.40, z-score = -2.14, R) >droEre2.scaffold_4690 4036094 69 - 18748788 UCACUGACUAGACUAGAGCUUGAGGUUCUCUGGG--GUAGUCUU--AACUAGGGAAUCCUCAGUCGCUCUGCA- .............((((((((((((((((((((.--........--..)))))))).))))))..))))))..- ( -24.60, z-score = -1.50, R) >consensus _______UGAGAUUAGAGCUUGAGGUUCUCUUGGGUAUAGUCUU_AAAGUAGGGAAUCCUCAGUCGCUCUAUCA .............((((((((((((((((((...................)))))).))))))..))))))... (-17.33 = -17.81 + 0.48)

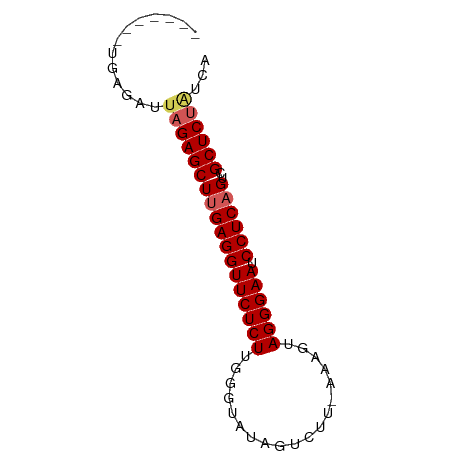
| Location | 12,129,770 – 12,129,836 |
|---|---|
| Length | 66 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.32375 |
| G+C content | 0.45504 |
| Mean single sequence MFE | -16.22 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12129770 66 - 22422827 UGAUAGAGCGACUGAGGAUUCCCUACUCU-AAGACAAUUCCCAAGAGAACCUCAAGCUCAAAUCUCA------- .....((((...(((((.(((.((.....-.............)).)))))))).))))........------- ( -14.27, z-score = -1.55, R) >droSim1.chrX 9351360 67 - 17042790 UGAUAGAGCGACUGAGGAUUCCCUACUUUCAAGACUAUAUCCAAGAGAACCUCAAGCUCUAAUCUCA------- .((((((((...(((((.(((.((...................)).)))))))).))))).)))...------- ( -16.61, z-score = -2.11, R) >droSec1.super_45 147769 67 - 245362 UGAUAGAGCGACUGAGGAUUCCCUACUUUCAAGACUAUACCCAAGAGAACCUCAAGCUCUAAUCUCA------- .((((((((...(((((.(((.((...................)).)))))))).))))).)))...------- ( -16.61, z-score = -2.53, R) >droYak2.chrX 20039125 65 + 21770863 UGCUAGAGCGACUGAGGAUUCCCUGGCU--AAGACUACAACCAAGAGAACCUCUAGCUCUUGUCUCA------- .((.(((((....((((.((((.(((..--..........))).).)))))))..))))).))....------- ( -17.70, z-score = -1.10, R) >droEre2.scaffold_4690 4036094 69 + 18748788 -UGCAGAGCGACUGAGGAUUCCCUAGUU--AAGACUAC--CCCAGAGAACCUCAAGCUCUAGUCUAGUCAGUGA -.(((((((...(((((.(((.(((((.--...)))..--...)).)))))))).))))).))........... ( -15.90, z-score = -0.18, R) >consensus UGAUAGAGCGACUGAGGAUUCCCUACUUU_AAGACUAUACCCAAGAGAACCUCAAGCUCUAAUCUCA_______ .((((((((...(((((.(((.((...................)).)))))))).))))).))).......... (-14.19 = -14.47 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:43 2011