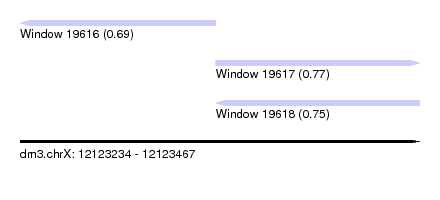
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,123,234 – 12,123,467 |
| Length | 233 |
| Max. P | 0.770254 |

| Location | 12,123,234 – 12,123,348 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 65.23 |
| Shannon entropy | 0.68388 |
| G+C content | 0.38985 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -6.43 |
| Energy contribution | -7.74 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12123234 114 - 22422827 AUUCCAACAAUUUCC-CUAAUUUCCCACAUUUGCCAGCGGCC----GGCAAAACACUCUUAAU---ACCGCUUAGCAAAUGUUAAUUGUUUUUCCCCUGGUAAGAACGAUUGCAUUUGCCUA ...............-.............((((((.......----))))))...........---........((((((((.((((((((((((...)).))))))))))))))))))... ( -24.80, z-score = -2.45, R) >droEre2.scaffold_4690 4030007 114 + 18748788 AUUCAAACAUUUUCC-CUAAUUUCCCACAUUUGCCAGCGACU----GGCAAAACUCUCCUAAU---ACCGCUUAGCAAAUGUUAAUUGUUUUUCCCCUGAUAACAACGAUUGCAUUUGCGUA ...............-..........((.((((((((...))----))))))...........---........((((((((.(((((((..((....))....))))))))))))))))). ( -23.60, z-score = -3.83, R) >droYak2.chrX 20033053 121 + 21770863 AUUCUAACAUUUCCC-CUAAUUUCCCACAUUUUCCAGCGGCCAACUGGCAAAACACUCUUAAUACCACCACUUAGCAAAUGUUAAUUGUUUUUCCCCUGGUAACAACGAUUGCAUUUCCCUA ...............-....................((((((....))).............(((((......(((((.......))))).......)))))........)))......... ( -13.22, z-score = -0.01, R) >droSec1.super_45 141359 114 - 245362 AUUCCAACAUUUUCC-CUAAUUUCCCACAUUUGCCAGCGGCU----GGCAAAACACUCUUAAU---ACCGCUUAGCAAAUGUUAAUUGUUUUUCCCCUGGUAACAACGAUUGCAUUUGCCUA ...............-.............((((((((...))----))))))...........---........((((((((.(((((((.((((...)).)).)))))))))))))))... ( -24.10, z-score = -2.51, R) >droSim1.chrX 9344972 114 - 17042790 AUUGCAACAUUUUCC-CUAAUUUCCCACAUUUGCCAGCGGCU----GGCAAAACACUCUUAAU---ACCGCUUAGCAAAUGUUAAUUGUUUUUCCCCUGGUAACAACGAUUGCAUUUGCCUA ...............-.............((((((((...))----))))))...........---........((((((((.(((((((.((((...)).)).)))))))))))))))... ( -24.10, z-score = -1.78, R) >droPer1.super_12 1956858 110 + 2414086 AUUGCCGCAUUUUCCACCUAUUUUUGAUCAUU--CUGUAACC----CUAAUCCCAGUCUUCGG---AUUUCUCACCGAGCG---AUCGAUCCCGAUUGGUCGAUCACGAUCACGAGUGUCCA ..................(((((.(((((...--........----.............((((---........))))(.(---(((((((......)))))))).)))))).))))).... ( -23.60, z-score = -0.89, R) >droGri2.scaffold_14624 3996128 118 + 4233967 UCUAUCGCAAACUUAACCUAAUCUAAGCCAAUAUUAUCGAUA----AUCAUAUCAGUAAAAACAAAAUAAUAUUGUGAGUGUUGGUUGAAUCCAUAAUAUUAAAAUAUAUGCCACAUUUUUA ......(((..((((........))))..((((((((.(((.----.(((.(((((((...((((.......))))...))))))))))))).))))))))........))).......... ( -14.80, z-score = -0.65, R) >consensus AUUCCAACAUUUUCC_CUAAUUUCCCACAUUUGCCAGCGGCC____GGCAAAACACUCUUAAU___ACCGCUUAGCAAAUGUUAAUUGUUUUUCCCCUGGUAACAACGAUUGCAUUUGCCUA ..........................................................................((((((((.(((((((.((.(....).)).)))))))))))))))... ( -6.43 = -7.74 + 1.31)



| Location | 12,123,348 – 12,123,467 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Shannon entropy | 0.03087 |
| G+C content | 0.26195 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12123348 119 + 22422827 UUUUGCUUUUAAUUACACGCUUGAUUAAGUGUUUACUUAAAUUGCGAAUUGAAAAGGUCGAUUUACAUGUUUCGGAAAUCCGCAUUAAUUCGACUUUAAUUAUAAACAAUAGAAUAAAU .(((((.(((((.((.((((((....)))))).)).)))))..))))).....((((((((.(((.(((...(((....))))))))).))))))))...................... ( -24.70, z-score = -2.45, R) >droSec1.super_45 141473 118 + 245362 UUUUGCUUUUAAUUACACGCUUGAUUAAGUGUUUACUUAAAUUGCGAAU-GAAAAGGUCGAUUUACAUGUUUAGCAAAUCCGUAUUAAUUCGACUUUAAUUAUAAACAAUAGAAUAAAU .(((((.(((((.((.((((((....)))))).)).)))))..))))).-...((((((((.(((.(((..(.....)..)))..))).))))))))...................... ( -19.60, z-score = -0.96, R) >droSim1.chrX 9345086 118 + 17042790 UUUUGCUUUUAAUUACACGCUUGAUUAAGUGUUUACUUAAAUUGCGAAU-GAAAAGGUCGAUUUACAUGUUUAGCAAAUCCGCAUUAAUUCGACUUUAAUUAUAAACAAUAGAAUAAAU .(((((.(((((.((.((((((....)))))).)).)))))..))))).-...((((((((.(((.((((...........))))))).))))))))...................... ( -21.70, z-score = -1.60, R) >consensus UUUUGCUUUUAAUUACACGCUUGAUUAAGUGUUUACUUAAAUUGCGAAU_GAAAAGGUCGAUUUACAUGUUUAGCAAAUCCGCAUUAAUUCGACUUUAAUUAUAAACAAUAGAAUAAAU .(((((.(((((.((.((((((....)))))).)).)))))..))))).....((((((((.(((.(((.............)))))).))))))))...................... (-20.37 = -20.15 + -0.22)


| Location | 12,123,348 – 12,123,467 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Shannon entropy | 0.03087 |
| G+C content | 0.26195 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12123348 119 - 22422827 AUUUAUUCUAUUGUUUAUAAUUAAAGUCGAAUUAAUGCGGAUUUCCGAAACAUGUAAAUCGACCUUUUCAAUUCGCAAUUUAAGUAAACACUUAAUCAAGCGUGUAAUUAAAAGCAAAA ..........((((((.((((((..(((((.(((((((((....)))...))).))).)))))..........(((...((((((....))))))....)))..)))))).)))))).. ( -23.40, z-score = -2.71, R) >droSec1.super_45 141473 118 - 245362 AUUUAUUCUAUUGUUUAUAAUUAAAGUCGAAUUAAUACGGAUUUGCUAAACAUGUAAAUCGACCUUUUC-AUUCGCAAUUUAAGUAAACACUUAAUCAAGCGUGUAAUUAAAAGCAAAA ..........((((((.((((((..((((........).(((((((.......))))))))))......-...(((...((((((....))))))....)))..)))))).)))))).. ( -20.30, z-score = -1.89, R) >droSim1.chrX 9345086 118 - 17042790 AUUUAUUCUAUUGUUUAUAAUUAAAGUCGAAUUAAUGCGGAUUUGCUAAACAUGUAAAUCGACCUUUUC-AUUCGCAAUUUAAGUAAACACUUAAUCAAGCGUGUAAUUAAAAGCAAAA ..........((((((.((((((....((......(((((((((((.......)))))))((.....))-...))))..((((((....)))))).....))..)))))).)))))).. ( -21.90, z-score = -1.93, R) >consensus AUUUAUUCUAUUGUUUAUAAUUAAAGUCGAAUUAAUGCGGAUUUGCUAAACAUGUAAAUCGACCUUUUC_AUUCGCAAUUUAAGUAAACACUUAAUCAAGCGUGUAAUUAAAAGCAAAA ..........((((((.((((((..((((........).(((((((.......))))))))))..........(((...((((((....))))))....)))..)))))).)))))).. (-18.63 = -18.97 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:40 2011