
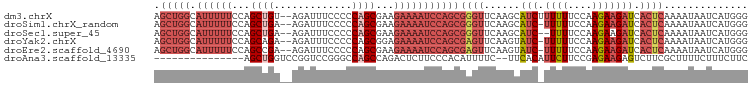
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,119,154 – 12,119,273 |
| Length | 119 |
| Max. P | 0.985546 |

| Location | 12,119,154 – 12,119,251 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Shannon entropy | 0.45672 |
| G+C content | 0.44649 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -14.81 |
| Energy contribution | -17.03 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
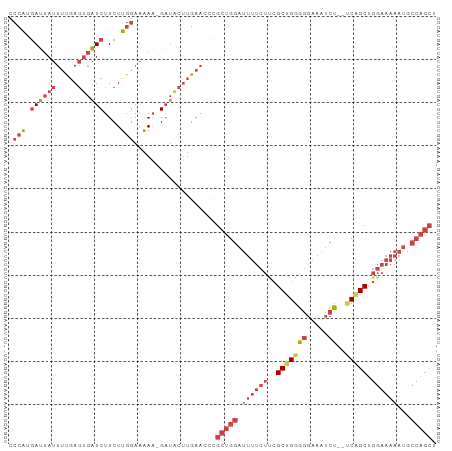
>dm3.chrX 12119154 97 + 22422827 CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAAAAGAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCU--ACAGCUGGAAAAAUGCCAGCU ...........((..(((.(((((.((....))))))))))..))...(((((.((((((..((((..((....))--.)))).))))))...))))). ( -27.40, z-score = -1.58, R) >droSim1.chrX_random 3301168 96 + 5698898 CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAAA-GAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCU--UCAGCUGGAAAAAUGCCAGCU ...........((..(((.(((((........))-))))))..))...(((((.((((((..(((((((.....))--))))).))))))...))))). ( -29.00, z-score = -1.69, R) >droSec1.super_45 137302 95 + 245362 CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAA--GAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCU--UCAGCUGGAAAAAUGCCAGCU ...........((..(((.(((((.......))--))))))..))...(((((.((((((..(((((((.....))--))))).))))))...))))). ( -28.70, z-score = -1.60, R) >droYak2.chrX 20029030 96 - 21770863 CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAAA-GAUACUUGAACUCGCUGGAUUUUCUCCGCUGGGGGAAAUCU--UCUGCUGGAAAAAUGCCAGCU ...........((..((((.((((........))-))))))..))...((((((((((.(((((.(((((....))--))))).)))))))).))))). ( -26.80, z-score = -1.77, R) >droEre2.scaffold_4690 4025994 96 - 18748788 CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAAA-GAUACUUGAACUCGCUGGAUUUUCUUCGCUGGGGGAAAUCU--UCGGCUGGAAAAAUGCCAGCU ...........((..((((.((((........))-))))))..))...(((((.((((((..(((((((.....))--))))).))))))...))))). ( -28.30, z-score = -2.05, R) >droAna3.scaffold_13335 2098330 82 - 3335858 GAAGAAAGAAAAGCGAAGACUCUUCUCGGAAGAAUGUGAA--GAAAAUGUGGGAAGAGUCUGGCUGGCCCGGACCGGACCAGCU--------------- ................((((((((((((.(..........--.....).))))))))))))((((((.(((...))).))))))--------------- ( -28.16, z-score = -2.98, R) >consensus CCCAUGAUUAUUUUGAGUGAUCUUCUUGGAAAAA_GAUACUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCU__UCAGCUGGAAAAAUGCCAGCU .(((.((((((.....))))))....)))...................(((((.((((((..((((..((....))...)))).))))))...))))). (-14.81 = -17.03 + 2.23)

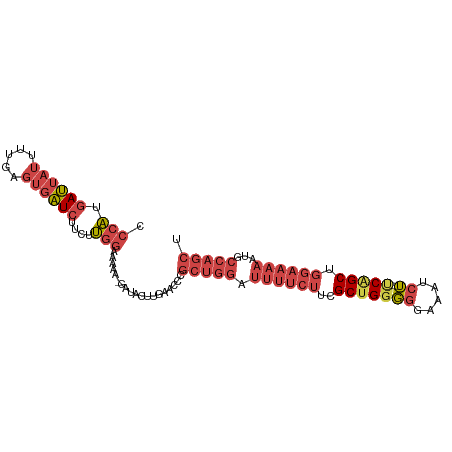
| Location | 12,119,154 – 12,119,251 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Shannon entropy | 0.45672 |
| G+C content | 0.44649 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -11.56 |
| Energy contribution | -13.37 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12119154 97 - 22422827 AGCUGGCAUUUUUCCAGCUGU--AGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUCUUUUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.((((((...((((.--.(.....)..))))...)))))))))))((((....(.(((((((.....)))))))))))).............. ( -23.80, z-score = -1.41, R) >droSim1.chrX_random 3301168 96 - 5698898 AGCUGGCAUUUUUCCAGCUGA--AGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUC-UUUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.((((((...((((.--..........))))...)))))))))))((((....(.(((-((((....)))))))))))).............. ( -22.80, z-score = -0.85, R) >droSec1.super_45 137302 95 - 245362 AGCUGGCAUUUUUCCAGCUGA--AGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUC--UUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.((((((...((((.--..........))))...)))))))))))((((....(.(((--(((.....))))))))))).............. ( -22.50, z-score = -0.81, R) >droYak2.chrX 20029030 96 + 21770863 AGCUGGCAUUUUUCCAGCAGA--AGAUUUCCCCCAGCGGAGAAAAUCCAGCGAGUUCAAGUAUC-UUUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.......)))))...--........(((((((((.....))).))((((....(.(((-((((....))))))))))))..........)))) ( -23.80, z-score = -2.02, R) >droEre2.scaffold_4690 4025994 96 + 18748788 AGCUGGCAUUUUUCCAGCCGA--AGAUUUCCCCCAGCGAAGAAAAUCCAGCGAGUUCAAGUAUC-UUUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.((((((...((.(.--..........).))...)))))))))))((((....(.(((-((((....)))))))))))).............. ( -19.10, z-score = -0.82, R) >droAna3.scaffold_13335 2098330 82 + 3335858 ---------------AGCUGGUCCGGUCCGGGCCAGCCAGACUCUUCCCACAUUUUC--UUCACAUUCUUCCGAGAAGAGUCUUCGCUUUUCUUUCUUC ---------------.(((((((((...))))))))).(((((((((.(........--.............).)))))))))................ ( -26.30, z-score = -3.89, R) >consensus AGCUGGCAUUUUUCCAGCUGA__AGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUC_UUUUUCCAAGAAGAUCACUCAAAAUAAUCAUGGG .(((((.((((((...((((.............))))...)))))))))))...................(((.((..((........))..)).))). (-11.56 = -13.37 + 1.81)
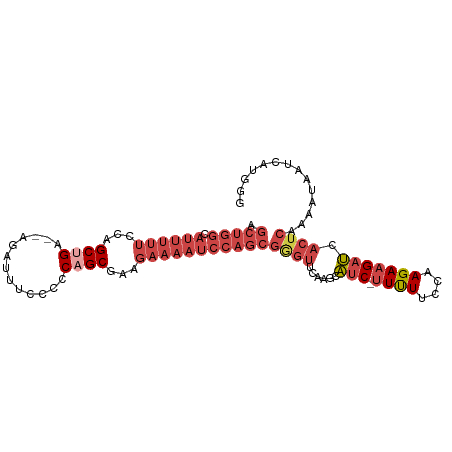
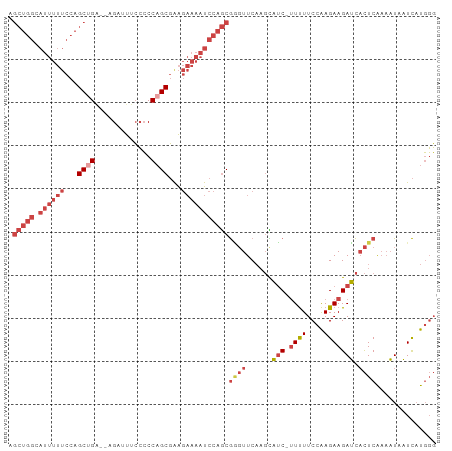
| Location | 12,119,181 – 12,119,273 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Shannon entropy | 0.45015 |
| G+C content | 0.46452 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -16.05 |
| Energy contribution | -17.50 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12119181 92 + 22422827 GGAAAAAAGAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCUACAGCUGGAAAAAUGCCAGCUGUU-UGAUGCAUCAUGAUCUUCU------- ........((((((..(((..(((((.((((((..((((..((....)).)))).))))))...))))).)))-..).)))))..........------- ( -31.30, z-score = -3.19, R) >droSim1.chrX_random 3301195 98 + 5698898 -GGAAAAAGAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCUUCAGCUGGAAAAAUGCCAGCUGUU-UGAUGCAUCAUGAUCCAUGAUCUUCU -(((..(.((((((..(((..(((((.((((((..(((((((.....))))))).))))))...))))).)))-..).))))).)..))).......... ( -36.30, z-score = -3.55, R) >droSec1.super_45 137329 97 + 245362 --GGAAAAGAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCUUCAGCUGGAAAAAUGCCAGCUGUU-UGAUGCAUCAUGAUCCAUGAUCUUCG --.(((..((((((..(((..(((((.((((((..(((((((.....))))))).))))))...))))).)))-..).)))))..((((...))))))). ( -35.80, z-score = -3.35, R) >droYak2.chrX 20029057 91 - 21770863 -GGAAAAAGAUACUUGAACUCGCUGGAUUUUCUCCGCUGGGGGAAAUCUUCUGCUGGAAAAAUGCCAGCUGCU-UGAUGCAUCAUGAUCUUCU------- -.....(((((...(((....((((((((((.(((((.(((((....))))))).)))))))).)))))(((.-....))))))..)))))..------- ( -27.00, z-score = -2.33, R) >droEre2.scaffold_4690 4026021 91 - 18748788 -GGAAAAAGAUACUUGAACUCGCUGGAUUUUCUUCGCUGGGGGAAAUCUUCGGCUGGAAAAAUGCCAGCUGUU-UGAUGCAUCAUGAUCUUCU------- -.......(((.((..(((..(((((.((((((..(((((((.....))))))).))))))...))))).)))-..).).)))..........------- ( -26.80, z-score = -2.21, R) >droAna3.scaffold_13335 2098357 78 - 3335858 -GGAAGAAUGUGAA-GAAAAUGUGGGAAGAGUCUGGCUGG-------CCCGGACCGGA------CCAGCUGCCAUGAUGGCUCAUGAUCUGUU------- -...(((.(((((.-...................((((((-------.(((...))).------))))))(((.....)))))))).)))...------- ( -21.80, z-score = -0.34, R) >consensus _GGAAAAAGAUGCUUGAACCCGCUGGAUUUUCUUCGCUGGGGGAAAUCUUCAGCUGGAAAAAUGCCAGCUGUU_UGAUGCAUCAUGAUCUUCU_______ ........(((...(((....(((((.((((((..(((((((.....))))))).))))))...)))))(((......))))))..)))........... (-16.05 = -17.50 + 1.45)


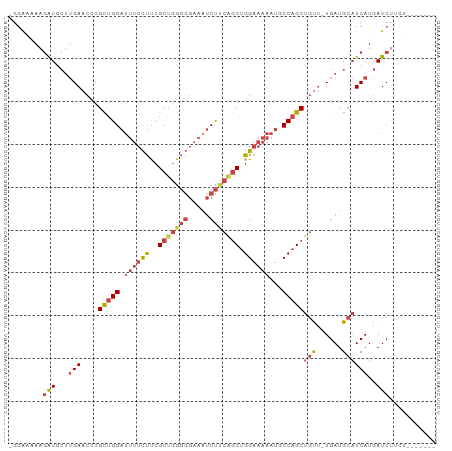
| Location | 12,119,181 – 12,119,273 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.31 |
| Shannon entropy | 0.45015 |
| G+C content | 0.46452 |
| Mean single sequence MFE | -25.31 |
| Consensus MFE | -12.13 |
| Energy contribution | -13.49 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12119181 92 - 22422827 -------AGAAGAUCAUGAUGCAUCA-AACAGCUGGCAUUUUUCCAGCUGUAGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUCUUUUUUCC -------.(((((....(((((....-(((.(((((.((((((...((((..(.....)..))))...)))))))))))..)))...)))))..))))). ( -28.70, z-score = -3.43, R) >droSim1.chrX_random 3301195 98 - 5698898 AGAAGAUCAUGGAUCAUGAUGCAUCA-AACAGCUGGCAUUUUUCCAGCUGAAGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUCUUUUUCC- ..........(((....(((((....-(((.(((((.((((((...((((...........))))...)))))))))))..)))...)))))....)))- ( -27.90, z-score = -1.81, R) >droSec1.super_45 137329 97 - 245362 CGAAGAUCAUGGAUCAUGAUGCAUCA-AACAGCUGGCAUUUUUCCAGCUGAAGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUCUUUUCC-- ..........(((....(((((....-(((.(((((.((((((...((((...........))))...)))))))))))..)))...)))))...)))-- ( -27.50, z-score = -1.68, R) >droYak2.chrX 20029057 91 + 21770863 -------AGAAGAUCAUGAUGCAUCA-AGCAGCUGGCAUUUUUCCAGCAGAAGAUUUCCCCCAGCGGAGAAAAUCCAGCGAGUUCAAGUAUCUUUUUCC- -------((((((((.((((((....-.)))(((((..(((((((.((...............)))))))))..)))))....))).).)))))))...- ( -23.96, z-score = -2.33, R) >droEre2.scaffold_4690 4026021 91 + 18748788 -------AGAAGAUCAUGAUGCAUCA-AACAGCUGGCAUUUUUCCAGCCGAAGAUUUCCCCCAGCGAAGAAAAUCCAGCGAGUUCAAGUAUCUUUUUCC- -------.(((((....(((((....-(((.(((((.((((((...((.(...........).))...)))))))))))..)))...))))).))))).- ( -20.20, z-score = -1.95, R) >droAna3.scaffold_13335 2098357 78 + 3335858 -------AACAGAUCAUGAGCCAUCAUGGCAGCUGG------UCCGGUCCGGG-------CCAGCCAGACUCUUCCCACAUUUUC-UUCACAUUCUUCC- -------...(((..(((.(((.....))).(((((------((((...))))-------))))).............)))..))-)............- ( -23.60, z-score = -2.54, R) >consensus _______AAAAGAUCAUGAUGCAUCA_AACAGCUGGCAUUUUUCCAGCUGAAGAUUUCCCCCAGCGAAGAAAAUCCAGCGGGUUCAAGCAUCUUUUUCC_ .................(((((.....(((.(((((.((((((...((...............))...)))))))))))..)))...)))))........ (-12.13 = -13.49 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:37 2011