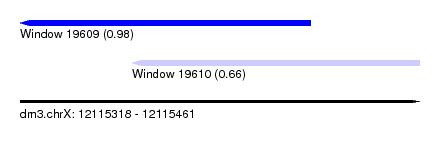
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,115,318 – 12,115,461 |
| Length | 143 |
| Max. P | 0.978587 |

| Location | 12,115,318 – 12,115,422 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.84 |
| Shannon entropy | 0.49848 |
| G+C content | 0.44513 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
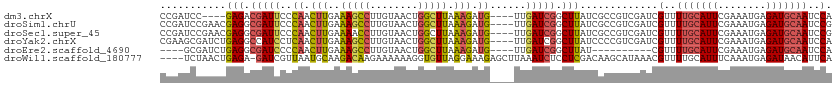
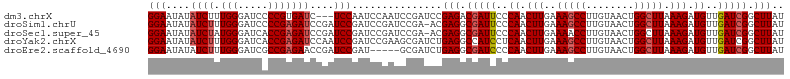
>dm3.chrX 12115318 104 - 22422827 CCGAUCC----GAGACGAUUCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUG----UUGAUCGGCUUAUCGCCGUCGAUCGUUUUGCAUUCGAAAUGAGAUGCAAUCCA ..((((.----..(((((((..((.(((..(((((........))))).))).))----..))))(((.....))))))))))(..(((((((........)))))))..). ( -32.70, z-score = -3.11, R) >droSim1.chrU 11937541 108 - 15797150 CCGAUCCGAACGAGGCGAUUCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUG----UUGAUCGGCUUAUCGCCGUCGAUCGUUUUGCAUUCGAAAUGAGAUGCAAUCCG .((((.(((..(((.(((((..((.(((..(((((........))))).))).))----..))))).))).)))..))))..((..(((((((........)))))))..)) ( -33.30, z-score = -2.28, R) >droSec1.super_45 133636 108 - 245362 CCGAUCCGAACGAGGCGAUUCCCAACUUGAAAACCUUGUAACUGGCUUAAAGAUG----UUGAUCGGCUUAUCGCCGUCGAUCGUUUUGCAUUCGAAAUGAGAUGCAAUCCG ..((((.((....((((((..((..........((........))......(((.----...)))))...)))))).))))))(..(((((((........)))))))..). ( -26.90, z-score = -0.66, R) >droYak2.chrX 20025230 108 + 21770863 CGAAGCGAUCUGAGGCCAUCCUCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUG----UUGAUCGGCUUAUCCCCGUCGAUCGUUUUGCAUUCGAAAUGAGAUGCAAUCCA ...((((...(((((....))))).(((..(((((........))))).))).))----))(((((((........)))))))(..(((((((........)))))))..). ( -31.90, z-score = -2.22, R) >droEre2.scaffold_4690 4022303 94 + 18748788 ----GCGAUCUGAGGCGAUCCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUG----UUGAUCGGCUUAU----------CGUUUUGCAUUCGAAAUGAGAUGCAAUCCA ----((.(((((((.(((((..((.(((..(((((........))))).))).))----..))))).))))(----------(((((((....)))))))))))))...... ( -29.80, z-score = -2.94, R) >droWil1.scaffold_180777 3873555 107 - 4753960 ----UCUAACUGAGA-GAUCGUUAAUGCAAGACAAGAAAAAAGGUGUUAGGAAAGAGCUUAAAUCUCCUCGACAAGCAUAAACGUUUUGCAUUUCAAAUGAGAUAACAUUCA ----......((..(-..(((((((((((((((...........((((((((..((.......)))))).)))).........))))))))))...)))))..)..)).... ( -18.45, z-score = -0.08, R) >consensus CCGAUCCAACUGAGGCGAUCCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUG____UUGAUCGGCUUAUCGCCGUCGAUCGUUUUGCAUUCGAAAUGAGAUGCAAUCCA ...........(((.(((((..((.(((..(((((........))))).))).))......))))).))).............(..(((((((........)))))))..). (-17.65 = -17.68 + 0.03)
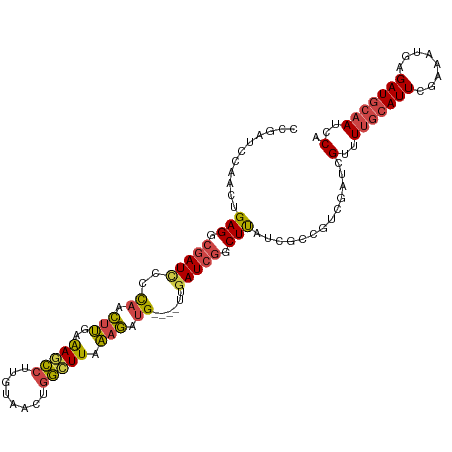
| Location | 12,115,358 – 12,115,461 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.52 |
| Shannon entropy | 0.18750 |
| G+C content | 0.46160 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12115358 103 - 22422827 GGAAUAUAUCUUUGGGAUCCCCGUGAUC---UCCAAUCCAAUCCGAUCCGAGACGAUUCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU (((........((((((((.....))))---.))))..........)))(((.(((((..((.(((..(((((........))))).))).))..))))).))).. ( -27.17, z-score = -1.59, R) >droSim1.chrU 11937581 105 - 15797150 GGAAUAUAUCUUUGGGAUCCCCGAGAUCCGAUCCGAUCCGAUCCGA-ACGAGGCGAUUCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU (((....(((..(.(((((.....))))).)...)))....)))..-..(((.(((((..((.(((..(((((........))))).))).))..))))).))).. ( -30.70, z-score = -1.39, R) >droSec1.super_45 133676 105 - 245362 GGAAUAUAUCUAUGGGAUCACCGAGAUCCGAUCCGAUCCGAUCCGA-ACGAGGCGAUUCCCAACUUGAAAACCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU (((....(((.((.(((((.....))))).))..)))....)))..-..(((.(((((..((.(((.....((........))....))).))..))))).))).. ( -25.20, z-score = -0.19, R) >droYak2.chrX 20025270 106 + 21770863 GGAAUAUAUCUUUGGGAUCACCGAGAUCCAAUCCGAUCCGAAGCGAUCUGAGGCCAUCCUCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU (((....(((((.((.....)))))))....)))((((......))))..((((((((..((.(((..(((((........))))).))).))..))).))))).. ( -30.40, z-score = -1.53, R) >droEre2.scaffold_4690 4022333 101 + 18748788 GGAAUAUAUCUUUGGGAUCGCCGAGAACCGAUCCGAU-----GCGAUCUGAGGCGAUCCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU .((..(((((((((((((((((.....(.((((....-----..)))).).)))))))))........(((((........)))))))))))))...))....... ( -33.80, z-score = -2.99, R) >consensus GGAAUAUAUCUUUGGGAUCACCGAGAUCCGAUCCGAUCCGAUCCGAUCCGAGGCGAUUCCCAACUUGAAAGCCUUGUAACUGGCUUAAAGAUGUUGAUCGGCUUAU (((....((((.(((.....)))))))....)))...............(((.(((((..((.(((..(((((........))))).))).))..))))).))).. (-24.84 = -25.40 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:34 2011