
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,143,465 – 11,143,585 |
| Length | 120 |
| Max. P | 0.783581 |

| Location | 11,143,465 – 11,143,585 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 145 |
| Reading direction | forward |
| Mean pairwise identity | 65.82 |
| Shannon entropy | 0.66450 |
| G+C content | 0.41570 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -15.54 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11143465 120 + 23011544 ------------------------UUUAGUAAUAAUCCAUUUAGCUGGGUUUUAAUCAG-AGAGUUACCUUGGAAGCCGGCGCUAAAGUUCGAGCAAAGGUGUGAAAUGUACUUUUGUUUGAGACAAUUGGAUCGGAGCAUUUCU ------------------------....((..(.((((((((((((.((((((.....(-((......))).)))))).).))))))(((((((((((((((.......))))))))))))).))...))))).)..))...... ( -32.90, z-score = -1.82, R) >droSim1.chr2L 10943274 116 + 22036055 ------------------------UUUUGUA-UAA---UGUUAACUGGGAUUUAAUUAG-GAAGUUACCUUGGAAGCCGGCGCUAAGGUUCUUGCAAAGGUGUGAAAUGUACUUUUGUUCGAGACAAUUGGAUCGGAGCAUUUCU ------------------------(((((((-.((---(.(((.((((..(((....((-(......)))..))).))))...))).)))..)))))))....((((((..(((..((((((.....))))))..))))))))). ( -23.80, z-score = 0.50, R) >droSec1.super_3 6544801 117 + 7220098 ------------------------UUUUGUAAUAA---GGUUAACUGGGAUUUUAUUAG-GAAGUUACCUUGGAAGCCGGCGCUAAGGUUCUUGCAAAGGUGUGAAAUGUACUUUUGUUUGAGACAAUUGGAUCGGAGCAUUUCU ------------------------((((.((((((---((((......)))))))))).-)))).......((((.((((..((((..(((..(((((((((.......)))))))))..)))....)))).))))....)))). ( -27.00, z-score = -0.42, R) >droYak2.chr2L 7539740 117 + 22324452 ------------------------UUUUGUAAUAA---AGUUGGUUGGGAUUUAAUCAG-GAACUUACCUUGGAAGCAGGAGCUAAGGUUCGUGCAAAGGUGUGAAAUGUGCUUUUGUUUGAGACAAUUGGAUCGGAGCAUUUCU ------------------------...........---..(((((((.....)))))))-.....((((((....(((.((((....)))).))).)))))).(((..((((((..(((..(.....)..)))..))))))))). ( -28.90, z-score = -1.00, R) >droEre2.scaffold_4929 12341145 117 - 26641161 ------------------------UUUUGUAAUGC---AGUUGGGUAGGAUAUAAUCAA-GAACUCACCUUGGAAGCUGGAGCCAAAGUUCGUGCAAAGGUGUGAAAUGUACUCUUGUUGGAGACAAUUGGAUCGGAGCAUUUCU ------------------------(((((((...(---((((((((((...........-...)).))))....)))))((((....)))).)))))))....(((((((.((((.((((....)))).)))...).))))))). ( -27.04, z-score = 0.31, R) >droAna3.scaffold_12916 11874965 98 - 16180835 ------------------------------------------UUUUGCAAAUUAAU-----ACUCUACCUUUGAGGCGGGGGCCAGAGUUCGAGCAAAGGUAUGAAAUGUACUCUUAUUGGAGACUAUGGAGUCGGCGCAUUUCU ------------------------------------------..............-----....((((((((.(((....))).(....)...)))))))).((((((((((((....(....)...)))))....))))))). ( -25.80, z-score = -0.74, R) >dp4.chr4_group3 6495588 112 + 11692001 ---------------------UUAAGGAGUU--GCUUGAGGUCUUUGAGAGGCAU----------UACCUUCGAGGCUGGGGCCAGAGUUCGUGCAAAGGUGGGAAAUGUGCUUUUGUUGGAGACAAUUGAGUCAGAGCAUUUCU ---------------------........((--((.((((.(((((((.(((...----------..)))))))(((....)))))).)))).)))).....((((((((((((..((((....)))).))))....)))))))) ( -33.50, z-score = -0.68, R) >droPer1.super_1 7987211 112 + 10282868 ---------------------UUAAGGAGUU--GCUUGAGGUCUUUGAGAGGCAU----------UACCUUCGAGGCUGGGGCCAGAGUUCGUGCAAAGGUGGGAAAUGUGCUUUUGUUGGAGACAAUUGAGUCAGAGCAUUUCU ---------------------........((--((.((((.(((((((.(((...----------..)))))))(((....)))))).)))).)))).....((((((((((((..((((....)))).))))....)))))))) ( -33.50, z-score = -0.68, R) >droWil1.scaffold_180708 7910856 138 - 12563649 AUGUGUGAGUGAGUGUGUUGUAUAGCAACGUAAACUUGAAUGCAUUUA-UAUUAAG------AAUUACCUUCGAGGCAGGUGCAAGUGUUCUUGCAUAGGUAUGAAACGUGCUCUUGUUGGAGACAAUUGAGUCUGAGCAUUUCU ......((((...(((((((.....))))))).))))((((((.((((-((((...------.....((.....))...(((((((....))))))).)))))))).((.((((..((((....)))).)))).)).)))))).. ( -36.20, z-score = -0.75, R) >droVir3.scaffold_12963 17857038 108 - 20206255 ---------------------------------AUUAGAAAAUAUUGCAUAUAAAUU----GAUCUACCUUCGAAGCGGGGGCGAGGGUCCGUGCAAAGGUGGGAAAGGUACUCUUAUUGGAAACAAUUGAGUCAGAGCAUUUCU ---------------------------------...(((((....(((.........----..((((((((....(((.((((....)))).))).))))))))....(.((((..((((....)))).)))))...)))))))) ( -31.90, z-score = -2.63, R) >droMoj3.scaffold_6500 4756258 124 - 32352404 ---------------------UUUAUAAAUUAGGCUGGGUACCUUUAAGAACCCCCUUUCGAACCUACCUUCGAAGCUGGGGCGAGUGUCCGUGCAAAGGUGGGAAAUGUGGUCUUAUUGGAGACUAUUGAGUCGGAGCAUUUCU ---------------------............(((((((.(......).))))((.(((((.((((((((....((..((((....))))..)).)))))))).....(((((((....))))))))))))..)))))...... ( -35.10, z-score = 0.06, R) >droGri2.scaffold_15252 15156078 111 - 17193109 ---------------------------ACCAAUAAUU-AUUGCAUUUAAUAUAAAU------AUUUACCUUUGAGGCAGGGGCAAGGGUUCGUGCAAAGGUAUGAAAUGUACUCUUCUUGGAUACAAUCGAGUCUGAACAUUUCU ---------------------------..........-..................------...(((((((...(((.((((....)))).)))))))))).(((((((((((...(((....)))..))))....))))))). ( -23.80, z-score = -0.97, R) >consensus ________________________UUUAGUAAUAAU__AGUUCAUUGAGAAUUAAU_____AACUUACCUUCGAAGCUGGGGCCAGAGUUCGUGCAAAGGUGUGAAAUGUACUCUUGUUGGAGACAAUUGAGUCGGAGCAUUUCU .................................................................((((((....((..((((....))))..)).)))))).(((((((..(((.(((.((.....)).))).)))))))))). (-15.54 = -14.92 + -0.62)
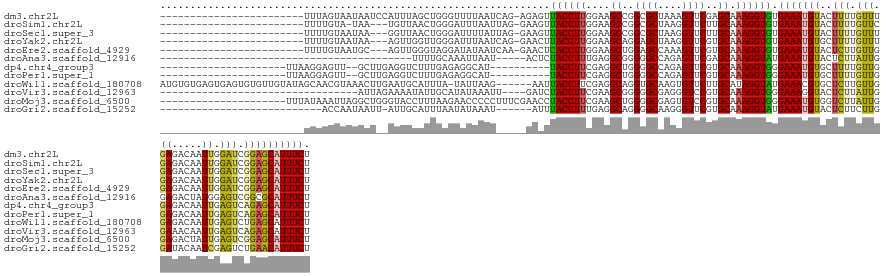
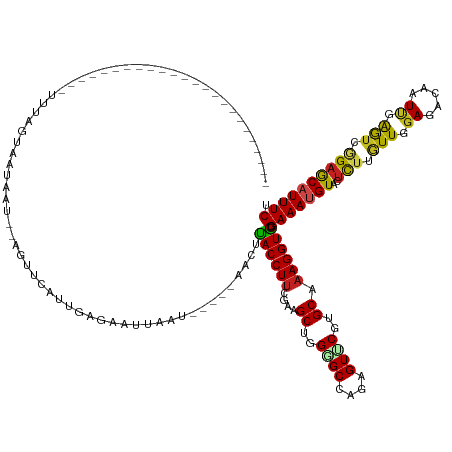
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:38 2011