
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,085,219 – 12,085,272 |
| Length | 53 |
| Max. P | 0.995613 |

| Location | 12,085,219 – 12,085,272 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Shannon entropy | 0.31252 |
| G+C content | 0.53525 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995613 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
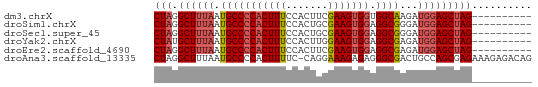
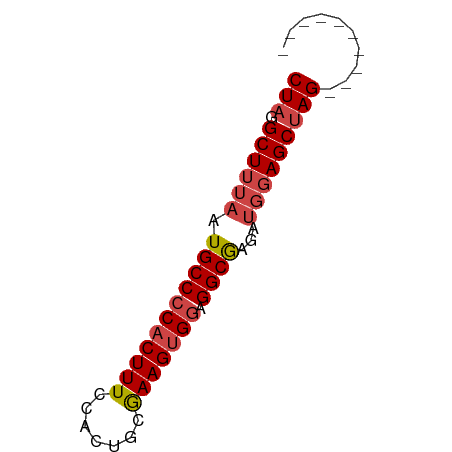
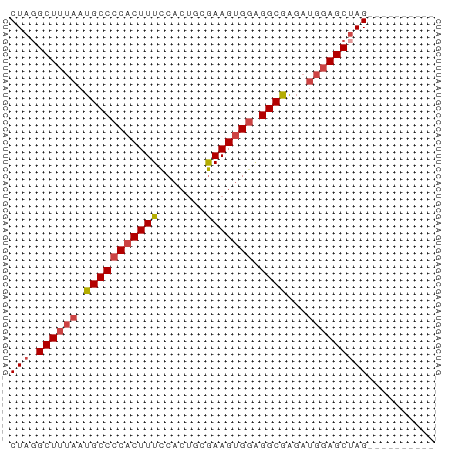
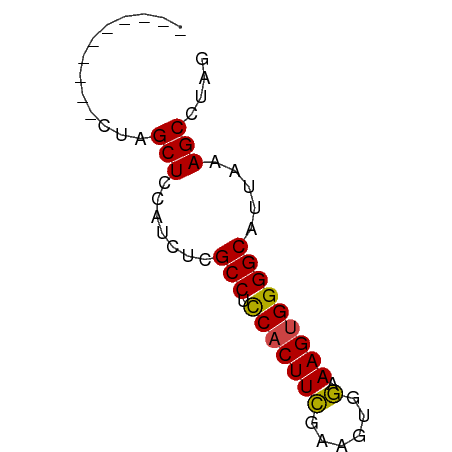
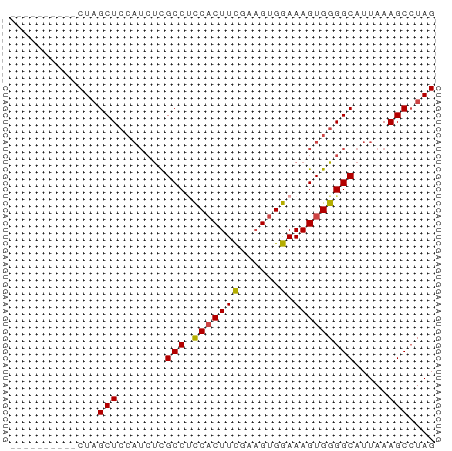
>dm3.chrX 12085219 53 + 22422827 CUAGGCUUUAAUGCCCCACUUUCCACUUCGAAGUGGUGGCAAGAUGGAGCUAG---------- ...(((((((.(((((((((((.......))))))).))))...)))))))..---------- ( -21.20, z-score = -3.31, R) >droSim1.chrX 9329358 53 + 17042790 CUAGGCUUUAAUGCCCCACUUUCCACUGCGAAGUGGAGGCGGGAUGGAGCUAG---------- ...(((((((.(((((((((((.......))))))).))))...)))))))..---------- ( -21.60, z-score = -2.54, R) >droSec1.super_45 104188 53 + 245362 CUAGGCUUUAAUGCCCCACUUUCCACUGCGAAGUGGAGGCGGGAUGGAGCUAG---------- ...(((((((.(((((((((((.......))))))).))))...)))))))..---------- ( -21.60, z-score = -2.54, R) >droYak2.chrX 19995610 53 - 21770863 CUAUGCUUUAAUGCCCCACUUUCCACUUGGAAGUGGAGGCGAGAUGGAGCUAG---------- (((.((((((.((((((((((.((....)))))))).))))...)))))))))---------- ( -22.20, z-score = -4.09, R) >droEre2.scaffold_4690 3991547 53 - 18748788 CUAGGCUUUAAUGCCCCACUUUCCACUUCGAAGUGGAGGCGAGAUGGAGCUAG---------- ...(((((((.(((((((((((.......))))))).))))...)))))))..---------- ( -20.50, z-score = -2.95, R) >droAna3.scaffold_13335 2054993 62 - 3335858 CUAGGCUUUAAUGCCCCACUUUUC-CAGGAAAGAGAGGGCGACUGCCAGCGAGAAAGAGACAG ...(((.....((((((.(((((.-...))))).).)))))...)))................ ( -17.10, z-score = -1.26, R) >consensus CUAGGCUUUAAUGCCCCACUUUCCACUGCGAAGUGGAGGCGAGAUGGAGCUAG__________ ....((((((.(((((((((((.......))))))).))))...))))))............. (-17.24 = -17.80 + 0.56)
| Location | 12,085,219 – 12,085,272 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Shannon entropy | 0.31252 |
| G+C content | 0.53525 |
| Mean single sequence MFE | -16.95 |
| Consensus MFE | -12.98 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921947 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
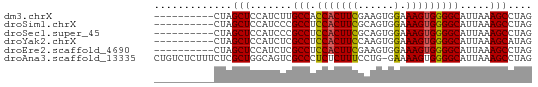
>dm3.chrX 12085219 53 - 22422827 ----------CUAGCUCCAUCUUGCCACCACUUCGAAGUGGAAAGUGGGGCAUUAAAGCCUAG ----------...((((((.(((....(((((....))))).)))))))))............ ( -17.20, z-score = -2.08, R) >droSim1.chrX 9329358 53 - 17042790 ----------CUAGCUCCAUCCCGCCUCCACUUCGCAGUGGAAAGUGGGGCAUUAAAGCCUAG ----------((((((...((((((.((((((....))))))..))))))......)).)))) ( -17.40, z-score = -1.94, R) >droSec1.super_45 104188 53 - 245362 ----------CUAGCUCCAUCCCGCCUCCACUUCGCAGUGGAAAGUGGGGCAUUAAAGCCUAG ----------((((((...((((((.((((((....))))))..))))))......)).)))) ( -17.40, z-score = -1.94, R) >droYak2.chrX 19995610 53 + 21770863 ----------CUAGCUCCAUCUCGCCUCCACUUCCAAGUGGAAAGUGGGGCAUUAAAGCAUAG ----------...(((.......(((.((((((((....)).))))))))).....))).... ( -17.90, z-score = -3.07, R) >droEre2.scaffold_4690 3991547 53 + 18748788 ----------CUAGCUCCAUCUCGCCUCCACUUCGAAGUGGAAAGUGGGGCAUUAAAGCCUAG ----------...((((((.((....((((((....)))))).))))))))............ ( -16.90, z-score = -2.04, R) >droAna3.scaffold_13335 2054993 62 + 3335858 CUGUCUCUUUCUCGCUGGCAGUCGCCCUCUCUUUCCUG-GAAAAGUGGGGCAUUAAAGCCUAG (((((...........)))))..(((((..((((....-..)))).)))))............ ( -14.90, z-score = -0.17, R) >consensus __________CUAGCUCCAUCUCGCCUCCACUUCGAAGUGGAAAGUGGGGCAUUAAAGCCUAG .............(((.......(((.(((((((......).))))))))).....))).... (-12.98 = -12.73 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:25 2011