| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,078,402 – 12,078,491 |
| Length | 89 |
| Max. P | 0.984922 |

| Location | 12,078,402 – 12,078,491 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 71.69 |
| Shannon entropy | 0.55671 |
| G+C content | 0.51479 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
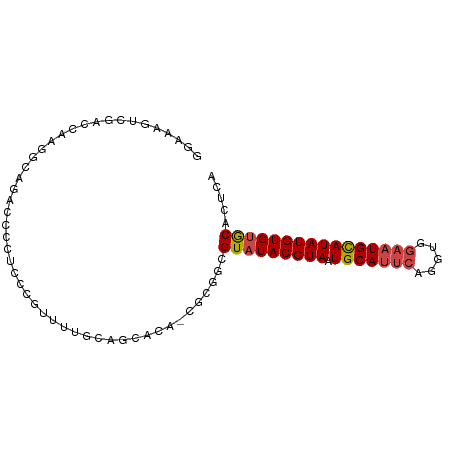
>dm3.chrX 12078402 89 - 22422827 GGAAAGUCGACCAAGGCAGAA--CCCCUUUUUUGCAGCACAGCGCGGCGUAUACGUAAUGCAUUCAGGUGGAAUGCAUAUGUGUGCACUCA .....((((......((((((--......)))))).((...)).))))(((((((((.(((((((.....))))))))))))))))..... ( -28.70, z-score = -1.31, R) >droSec1.super_45 98478 88 - 245362 GGAAAGUCGACCAAGGCAGAC--CUCCCUUUUUGGAGCACA-CGCGGCGUAUACGUAAUGCAUUCAGGUGGAAUGCAUAUGUGUGCACUCA .....(((..((((((.((..--....)))))))).((...-.)))))(((((((((.(((((((.....))))))))))))))))..... ( -28.50, z-score = -1.42, R) >droYak2.chrX 19988439 90 + 21770863 GGAAAACCGACCAAGGCAGACCCCUGCUGCAAAACAGCACA-CGCGACGUAUACGUAAUGCAUUCAGGUGGAAUGUAUAUGUGUGCACUCA ((....))......(((((....)))))........(((((-((..(((....))).((((((((.....)))))))).)))))))..... ( -26.50, z-score = -1.32, R) >droEre2.scaffold_4690 3986610 90 + 18748788 GGAAAGUCGACCACGGCGGACCCCUCCCGUUUUGCAGCGCA-CGCGGCGUAUACGUAAUGCAUUCAGGUGGAAUGCAUAUGUGUGCACUCA .....(((...((..((((.......))))..))..((...-.)))))(((((((((.(((((((.....))))))))))))))))..... ( -30.20, z-score = -0.32, R) >droAna3.scaffold_13335 2050109 89 + 3335858 GGUGGGCCAGAGAUCCCAAGGCAGGAUCUCGUUG-AGUGCA-CGCGGCGUAUACGUAAUGCAUUCAGGUGGAAUGCAUAUGUGUGCACUCA ((....)).(((((((.......)))))))..((-((((((-(((((((....))).((((((((.....)))))))).)))))))))))) ( -43.10, z-score = -4.79, R) >droGri2.scaffold_15081 150806 73 + 4274704 ----------GAGUACCAGAACCCAAACGAGACCAAACA---CGAGGCGUAUACGUAAUGCAUGCA-----AUUGCAUAUGUGCACACUCA ----------((((................(.((.....---...)))((.((((((.((((....-----..)))))))))).)))))). ( -15.40, z-score = -1.18, R) >consensus GGAAAGUCGACCAAGGCAGACCCCUCCCGUUUUGCAGCACA_CGCGGCGUAUACGUAAUGCAUUCAGGUGGAAUGCAUAUGUGUGCACUCA ....................................((.....))...(((((((((.(((((((.....))))))))))))))))..... (-17.25 = -17.67 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:22 2011