
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,138,087 – 11,138,164 |
| Length | 77 |
| Max. P | 0.696259 |

| Location | 11,138,087 – 11,138,164 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Shannon entropy | 0.49525 |
| G+C content | 0.49069 |
| Mean single sequence MFE | -14.90 |
| Consensus MFE | -7.96 |
| Energy contribution | -7.62 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11138087 77 - 23011544 CCAUCCUCCUUGGCCUCAUACUUGCCCAGAAUUAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAAAAAUAAAGAUGA-- .((((......(((((.((.((.....)).)).)))))...(((((.((....)))))))............)))).-- ( -17.70, z-score = -2.43, R) >anoGam1.chr3R 15587813 79 + 53272125 CCACCCUCCUUCGCCUCGUACCGGCCGAAAAUCAGGCCCAUAAAUUCGCUCAUGCAGUUACCUUGAAGAAGAAAAGUGG ((((....((((.....(((.(((((........)))).........((....)).).)))......))))....)))) ( -17.90, z-score = -1.50, R) >droGri2.scaffold_15252 15150492 75 + 17193109 CCUUCUUCCUUCGCCUCGUAGCGACCCAAUAUGAGUCCCAUGAAUUCGCUCAUGCAAUUUCCUGCACCACA-AUCC--- ..........((((......))))......((((((...........))))))(((......)))......-....--- ( -10.40, z-score = -1.48, R) >droMoj3.scaffold_6500 4750183 76 + 32352404 CCCUCCUCCUUGGCUUCAUAGCGACCGAGUAUGAGGCCCAUGAACUCGCUCAUGCAGUUUCCUUUAAGAUAGAUCU--- ........((((((((((((.(......))))))))))...(((((.((....))))))).....)))........--- ( -16.90, z-score = -0.49, R) >droVir3.scaffold_12963 17852081 73 + 20206255 CCCUCCUCCUUGGCUUCGUAGCGGCCGAGUAUGAGACCCAUGAAUUCGCUCAUGCAAUUCCCUUCAAGA---AUUU--- ..(((...(((((((.......)))))))...)))......(((((.((....))))))).........---....--- ( -15.90, z-score = -0.88, R) >dp4.chr4_group3 6489795 77 - 11692001 CCAUCCUCCUUGGCCUCAUAUUUACCCAAAAUCAAGCCCAUGAACUCGCUCAUGCAGUUUCCUAUCGAAAAAGUGCA-- ...........(((.....((((.....))))...))).((((......))))(((.((((.....))))...))).-- ( -6.20, z-score = 0.92, R) >droAna3.scaffold_12916 11869680 79 + 16180835 CCCUCCUCCUUGGCCUCGUACUUGCCCAGAAUGAGGCCCAUAAACUCACUCAUGCAGUUUCCUAGUUUAAUAAAUAAAA ...........((((((((.((.....)).))))))))..((((((.(((.....))).....)))))).......... ( -15.00, z-score = -2.02, R) >droEre2.scaffold_4929 12335857 77 + 26641161 CCAUCCUCCUUGGCCUCAUACCUGCCCAGAAUCAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAACAACACAGAUAG-- ..(((......(((((.((..((....)).)).)))))...(((((.((....)))))))............)))..-- ( -13.90, z-score = -1.18, R) >droYak2.chr2L 7534401 77 - 22324452 CCAUCCUCCUUGGCCUCAUACUUGCCCAGAAUCAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAAAAAUAAAGACAA-- ........((((((((.((.((.....)).)).)))))...(((((.((....)))))))..........)))....-- ( -14.60, z-score = -1.65, R) >droSec1.super_3 6539477 77 - 7220098 CCAUCCUCCUUGGCCUCAUACUUGCCCAGAAUCAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAACAAUAAAGAUGA-- .((((......(((((.((.((.....)).)).)))))...(((((.((....)))))))............)))).-- ( -17.70, z-score = -2.15, R) >droSim1.chr2L 10937940 77 - 22036055 CCAUCCUCCUUGGCCUCAUACUUGCCCAGAAUCAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAACAAUAAAGAUGA-- .((((......(((((.((.((.....)).)).)))))...(((((.((....)))))))............)))).-- ( -17.70, z-score = -2.15, R) >consensus CCAUCCUCCUUGGCCUCAUACUUGCCCAGAAUCAGGCCCAUGAACUCGCUCAUGCAGUUCCCUAAAAAAAAAGAUGA__ ...........(((((.((...........)).)))))...(((((.((....)))))))................... ( -7.96 = -7.62 + -0.35)

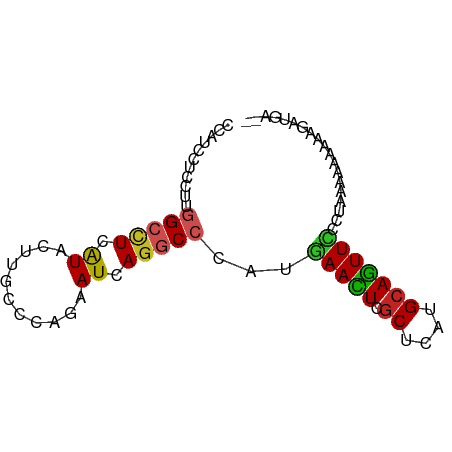
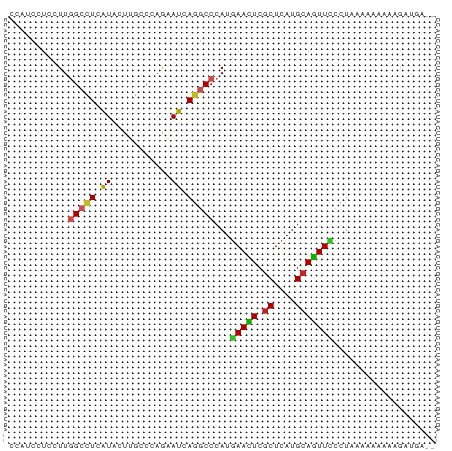
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:37 2011