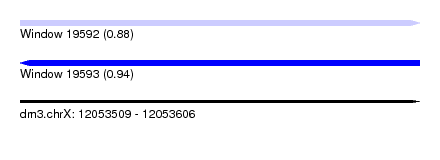
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,053,509 – 12,053,606 |
| Length | 97 |
| Max. P | 0.942401 |

| Location | 12,053,509 – 12,053,606 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.95 |
| Shannon entropy | 0.60344 |
| G+C content | 0.50519 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -9.81 |
| Energy contribution | -11.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
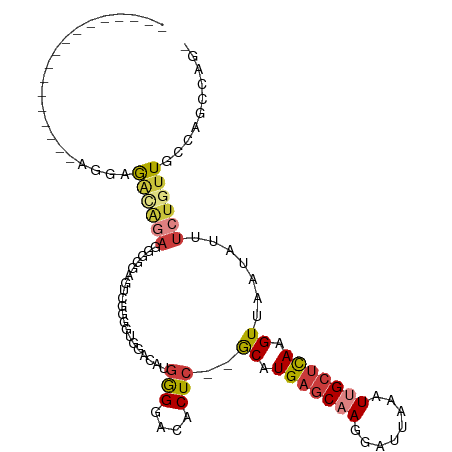
>dm3.chrX 12053509 97 + 22422827 ---------GGGGGGAUGGGGGGCAGAUGGGGAGUCGGGCU-GACAUGGGGACACUC--GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCAG- ---------....((...((.((((((.(((..(((.....-))).((....)))))--((.(((((((........))))))).)).......)))))).))..))..- ( -31.00, z-score = -1.68, R) >droSim1.chrX 9307143 90 + 17042790 -----------------UGGCAACAGAGGAGGAGUUGGGGCUGACAUGGGGACACUC--GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCAG- -----------------((((((((((((((..(((......))).((....)))))--((.(((((((........))))))).))......))))))))))).....- ( -33.50, z-score = -3.64, R) >droSec1.super_45 79310 90 + 245362 -----------------UGGCAACAGAGGAGGAGUUGGGGCUGACAUGGGGACACUC--GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCAG- -----------------((((((((((((((..(((......))).((....)))))--((.(((((((........))))))).))......))))))))))).....- ( -33.50, z-score = -3.64, R) >droYak2.chrX 19971075 86 - 21770863 --------------------UGGCAAACAGGGGCGCUGAA-GGCGAUGGGGACACUC--GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCAG- --------------------((((((.(((..(.......-.((((((....)).))--)).(((((((........)))))))........)..))))))))).....- ( -32.10, z-score = -3.23, R) >droEre2.scaffold_4690 3969176 89 - 18748788 --------------GGUAGGAGGGGGUUGGGGGGGGGGG----GCAUGGGGACACUC--GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCCG- --------------.........(((((((.((.((..(----((.((....))...--)).(((((((........)))))))........)..)).)).))))))).- ( -33.00, z-score = -3.18, R) >droAna3.scaffold_13335 2031883 99 - 3335858 -----------CUACUAACGAGAUAGAGAGAGAGAGGAAAUGGGUGCAAGGACCCUCCCACAUGAGCCAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUACCAGGCCGG -----------..........((((((((....(.(((...((((......))))))))((.(((((.(........).))))).)).....)))))))).......... ( -19.50, z-score = 0.77, R) >droPer1.super_12 1887339 105 - 2414086 AGCACACCAGGGCAUACCAGAGCACAACAGAGUCCACCGGUGGGAACGAG-AUGCUC--GCAUGCCCAAGGAUUAAAUCGCUUAAGUUAAUAUUUCUGUUACAAGGCA-- ......((.((((((....(((((.....(..((((....))))..)...-.)))))--..))))))..))........((((....(((((....)))))..)))).-- ( -25.50, z-score = -0.49, R) >consensus _________________AGGAGACAGAGGGGGAGUCGGGGUGGACAUGGGGACACUC__GCAUGAGCAAGGAUUAAAUUGCUCAAGUUAAUAUUUCUGUUGCCAGCCAG_ ..............................................(((.((((.....((.(((((((........))))))).)).........)))).)))...... ( -9.81 = -11.05 + 1.25)

| Location | 12,053,509 – 12,053,606 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.95 |
| Shannon entropy | 0.60344 |
| G+C content | 0.50519 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.64 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
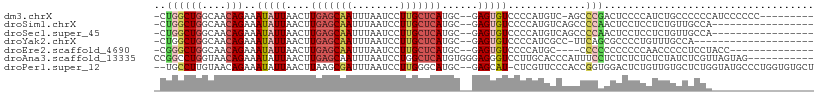
>dm3.chrX 12053509 97 - 22422827 -CUGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC--GAGUGUCCCCAUGUC-AGCCCGACUCCCCAUCUGCCCCCCAUCCCCCC--------- -..((((((((...(..(((((..(.(((((((........))))))).).--.)))))..)..))))-))))............................--------- ( -23.60, z-score = -3.01, R) >droSim1.chrX 9307143 90 - 17042790 -CUGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC--GAGUGUCCCCAUGUCAGCCCCAACUCCUCCUCUGUUGCCA----------------- -......(((((((((..........(((((((........)))))))...--((((..................))))....))))))))).----------------- ( -26.07, z-score = -3.39, R) >droSec1.super_45 79310 90 - 245362 -CUGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC--GAGUGUCCCCAUGUCAGCCCCAACUCCUCCUCUGUUGCCA----------------- -......(((((((((..........(((((((........)))))))...--((((..................))))....))))))))).----------------- ( -26.07, z-score = -3.39, R) >droYak2.chrX 19971075 86 + 21770863 -CUGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC--GAGUGUCCCCAUCGCC-UUCAGCGCCCCUGUUUGCCA-------------------- -......((((((((...........(((((((........))))))).((--((.((....)))))).-..........))).))))).-------------------- ( -22.60, z-score = -1.57, R) >droEre2.scaffold_4690 3969176 89 + 18748788 -CGGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC--GAGUGUCCCCAUGC----CCCCCCCCCCCAACCCCCUCCUACC-------------- -.(((..((((...(..(((((..(.(((((((........))))))).).--.)))))..)..)))----)..)))...................-------------- ( -21.10, z-score = -3.06, R) >droAna3.scaffold_13335 2031883 99 + 3335858 CCGGCCUGGUAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUGGCUCAUGUGGGAGGGUCCUUGCACCCAUUUCCUCUCUCUCUCUAUCUCGUUAGUAG----------- .....(((.(((((((.......((.(((((............))))).))(((((((..............)))))))........))).)))).)))----------- ( -21.64, z-score = 0.00, R) >droPer1.super_12 1887339 105 + 2414086 --UGCCUUGUAACAGAAAUAUUAACUUAAGCGAUUUAAUCCUUGGGCAUGC--GAGCAU-CUCGUUCCCACCGGUGGACUCUGUUGUGCUCUGGUAUGCCCUGGUGUGCU --..........................((((.......((..((((((((--((((((-..((...(((....)))....))..))))))..)))))))).))..)))) ( -28.80, z-score = -0.67, R) >consensus _CUGGCUGGCAACAGAAAUAUUAACUUGAGCAAUUUAAUCCUUGCUCAUGC__GAGUGUCCCCAUGCCCACCCCAACUCCCCCUCUGCCUCCA_________________ ..((((((....)))...........(((((((........)))))))........................)))................................... ( -9.07 = -9.64 + 0.57)
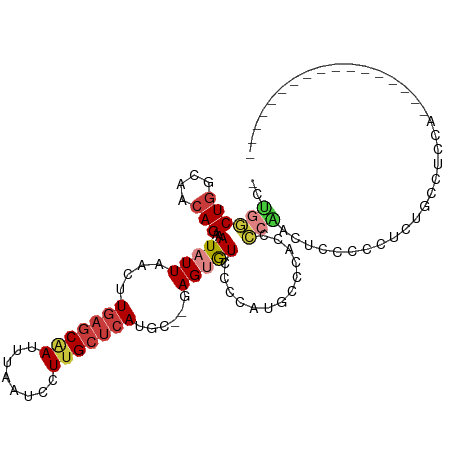
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:21 2011