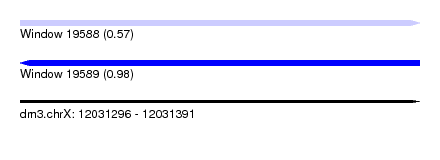
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,031,296 – 12,031,391 |
| Length | 95 |
| Max. P | 0.976843 |

| Location | 12,031,296 – 12,031,391 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Shannon entropy | 0.34343 |
| G+C content | 0.39955 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12031296 95 + 22422827 ACAGUUAACCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGAUGAUGGGGUUUUGGCAUCGUCAUUAUCAUUACCAUUAACAAAAGGCCACCG ...(((((..(((((((((((((..(((..(((.((....)).)))............)))..))))))))))).)).)))))............ ( -21.34, z-score = -0.54, R) >droSim1.chrX 9283870 92 + 17042790 ACAGUUAACCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGA---UGGGGUUUUGGCAUCGUCAUUAUCAUUACCAUUAACAAAAGGCCACCG ...(((((..(((((((((((((..(((..((.(((........---....))).)).)))..))))))))))).)).)))))............ ( -21.50, z-score = -0.94, R) >droSec1.super_45 57616 92 + 245362 ACAGUUACCCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGA---CGGGGUUUUGGCAUCGUCAUUAUGAUUACCAUUAACAAAAGGCCACCG ...(((((((...(((...((....))...)))....))).)))---)((((((((((.(((((....)))))..)))........))))).)). ( -18.60, z-score = 0.21, R) >droYak2.chrX 19949025 92 - 21770863 ACAGUUAGCCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGAU--GGGGUUUUUGGCAUCGUCAUUAUCAUUACCAUUAACAAAAG-CCAACG ...(((.((...((((.((((((((((((..(((.(((((((...--...))))))).)))..)))))))))))).)))).......)-).))). ( -22.50, z-score = -1.68, R) >droEre2.scaffold_4690 3948358 91 - 18748788 ACAGUUAACCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGA---UGGGGUUU-GGCAUCGUCAUUAUCAUUACCAUUAACAAAAGGCCACCG ...(((((..(((((((((((((..(((..(((.((....)).)---))......-..)))..))))))))))).)).)))))............ ( -20.60, z-score = -0.72, R) >droAna3.scaffold_13335 2008067 75 - 3335858 ---------CGAAAUGAUAAUGAUAAUGAAC----UGCGGCUUU---CACCGUAUCAUCAUUAUCAUUAUCGUAACCAUUAACCAAGGACC---- ---------....((((((((((((((((..----(((((....---..)))))...))))))))))))))))..................---- ( -24.70, z-score = -5.85, R) >consensus ACAGUUAACCGGAAUGAUAAUGAUAAUGAGCAUGAUAGGGAUGA___UGGGGUUUUGGCAUCGUCAUUAUCAUUACCAUUAACAAAAGGCCACCG ..........((((((((((((((.(((..............................))).)))))))))))).)).................. (-13.75 = -13.98 + 0.22)


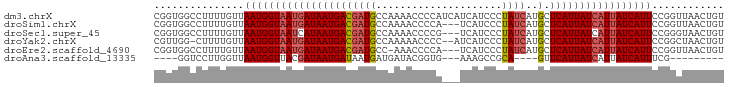
| Location | 12,031,296 – 12,031,391 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Shannon entropy | 0.34343 |
| G+C content | 0.39955 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -14.31 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12031296 95 - 22422827 CGGUGGCCUUUUGUUAAUGGUAAUGAUAAUGACGAUGCCAAAACCCCAUCAUCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGUUAACUGU ((((((((.......(((((((((((((((((.((((.........))))..(((.......))).)))))))))))))))))..)))).)))). ( -23.20, z-score = -2.07, R) >droSim1.chrX 9283870 92 - 17042790 CGGUGGCCUUUUGUUAAUGGUAAUGAUAAUGACGAUGCCAAAACCCCA---UCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGUUAACUGU ((((((((...((.((((((((((((..((((.((((...........---.))))....))))..)))))))))))).))....)))).)))). ( -22.90, z-score = -2.16, R) >droSec1.super_45 57616 92 - 245362 CGGUGGCCUUUUGUUAAUGGUAAUCAUAAUGACGAUGCCAAAACCCCG---UCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGGUAACUGU ((((.((((..((.((((((((((....((((.((((.(........)---.))))....))))....)))))))))).))....)))).)))). ( -21.60, z-score = -1.37, R) >droYak2.chrX 19949025 92 + 21770863 CGUUGG-CUUUUGUUAAUGGUAAUGAUAAUGACGAUGCCAAAAACCCC--AUCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGCUAACUGU .(((((-((......(((((((((((((((((.((((..........)--)))....(......).)))))))))))))))))..)))))))... ( -25.80, z-score = -4.07, R) >droEre2.scaffold_4690 3948358 91 + 18748788 CGGUGGCCUUUUGUUAAUGGUAAUGAUAAUGACGAUGCC-AAACCCCA---UCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGUUAACUGU ((((((((.......(((((((((((((((((.((((..-......))---))....(......).)))))))))))))))))..)))).)))). ( -23.40, z-score = -2.36, R) >droAna3.scaffold_13335 2008067 75 + 3335858 ----GGUCCUUGGUUAAUGGUUACGAUAAUGAUAAUGAUGAUACGGUG---AAAGCCGCA----GUUCAUUAUCAUUAUCAUUUCG--------- ----(..((.........))...)((((((((((((((.....((((.---...))))..----..))))))))))))))......--------- ( -24.80, z-score = -3.90, R) >consensus CGGUGGCCUUUUGUUAAUGGUAAUGAUAAUGACGAUGCCAAAACCCCA___UCAUCCCUAUCAUGCUCAUUAUCAUUAUCAUUCCGGUUAACUGU ...............(((((((((((((((((.((((.....................))))....)))))))))))))))))............ (-14.31 = -15.00 + 0.69)
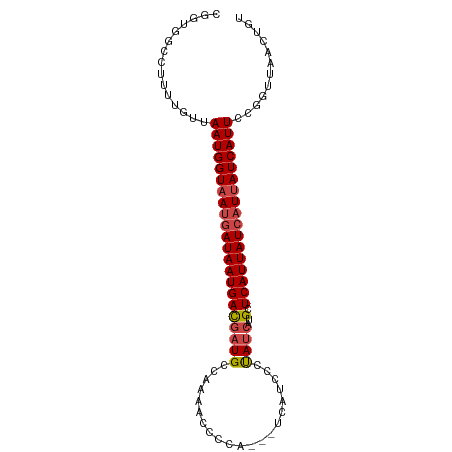
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:18 2011