
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,026,439 – 12,026,537 |
| Length | 98 |
| Max. P | 0.577234 |

| Location | 12,026,439 – 12,026,537 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.07 |
| Shannon entropy | 0.52105 |
| G+C content | 0.45461 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -11.27 |
| Energy contribution | -12.36 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
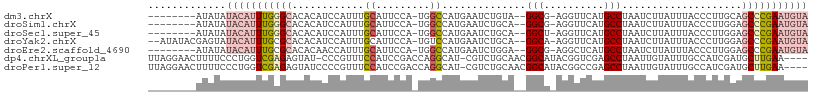
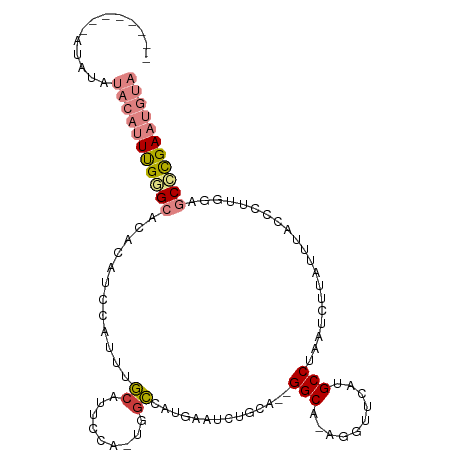
>dm3.chrX 12026439 98 - 22422827 --------AUAUAUACAUUUGGGCACACAUCCAUUUGCAUUCCA-UGGCCAUGAAUCUGUA--GGCG-AGGUUCAUGCCUAAUCUUAUUUACCCUUGCAGCCCGAAUGUA --------.....(((((((((((...((......)).......-.(((.(((((((((..--..).-)))))))))))....................))))))))))) ( -28.70, z-score = -2.19, R) >droSim1.chrX 9279110 98 - 17042790 --------AUAUAUACAUUUGGGCACACAUCCAUUUGCAUUCCA-UGGCCAUGAAUCUGCA--GGCG-AGGUUCAUGCCUAAUCUUAUUUACCCUUGGAGCCCGAAUGUA --------.....(((((((((((...((......))...((((-.(((.(((((((((..--..).-)))))))))))(((......)))....))))))))))))))) ( -30.90, z-score = -2.45, R) >droSec1.super_45 52533 98 - 245362 --------AUAUAUACAUUUGGGCACACAUCCAUUUGCAUUCCA-UGGCCAUGAAUCUGCA--GGCU-AGGUUCAUGCCUAAUCUUAUUUACCCUUGGAGCCCGAAUGUA --------.....(((((((((((...((......))...((((-.(((.(((((((((..--...)-)))))))))))(((......)))....))))))))))))))) ( -31.40, z-score = -2.77, R) >droYak2.chrX 19944565 104 + 21770863 --AUAUACGAGUAUACAUUUGCGCACACAUCCAUUUGCAUUCCA-UGUCCAUGAAUCUGCA--GGCA-AGGUUCAUGCCUAAUCUUAUUUACCCUUGGAGCCCGAAUGUA --...........((((((((.((.....((((...((((...)-))).(((((((((...--....-)))))))))..................)))))).)))))))) ( -21.10, z-score = -0.23, R) >droEre2.scaffold_4690 3943741 98 + 18748788 --------AUAUAUACAUUUGCGCACACAACCAUUUGCAUUCCA-UGGCCAUGAAUCUGGA--GGCG-AGGCUCAUGCCUAAUCUUAUUUACCCUUGGAGCCCGAAUGUA --------.....((((((((.((...(((....)))...((((-.((...(((((..(((--....-((((....))))..))).))))).)).)))))).)))))))) ( -24.90, z-score = -1.01, R) >dp4.chrXL_group1a 2749819 104 - 9151740 UUAGGAACUUUUCCCUGGUCGAGAGUAU-CCCGUUUCCAUCCGACCAGGCAU-CGUCUGCAACGGCAUACGGUCGAGCCUAAUUGUAUUUGCCAUCGAUGCUUGAA---- ...(((.....)))(((((((.((....-..........)))))))))((((-((...((((..(((...((.....))....)))..))))...)))))).....---- ( -30.74, z-score = -1.54, R) >droPer1.super_12 1853551 105 + 2414086 UUAGGAACUUUUCCCUGGUCGAGAGUAUCCCCGUUUCCAUCCGACCAGGCAU-CGUCUGCAACGGCAUACGGCCGAGCCUAAUUGUAUUUGCCAUCGAUGCUUGAA---- ...(((.....)))(((((((.((...............)))))))))((((-((...((((..(((...(((...)))....)))..))))...)))))).....---- ( -31.76, z-score = -1.82, R) >consensus ________AUAUAUACAUUUGGGCACACAUCCAUUUGCAUUCCA_UGGCCAUGAAUCUGCA__GGCA_AGGUUCAUGCCUAAUCUUAUUUACCCUUGGAGCCCGAAUGUA .............(((((((((((............((.........))..............(((..........)))....................))))))))))) (-11.27 = -12.36 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:15 2011