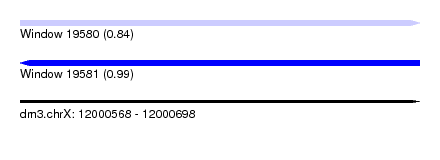
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,000,568 – 12,000,698 |
| Length | 130 |
| Max. P | 0.990708 |

| Location | 12,000,568 – 12,000,698 |
|---|---|
| Length | 130 |
| Sequences | 12 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Shannon entropy | 0.29754 |
| G+C content | 0.49643 |
| Mean single sequence MFE | -46.96 |
| Consensus MFE | -35.17 |
| Energy contribution | -35.15 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
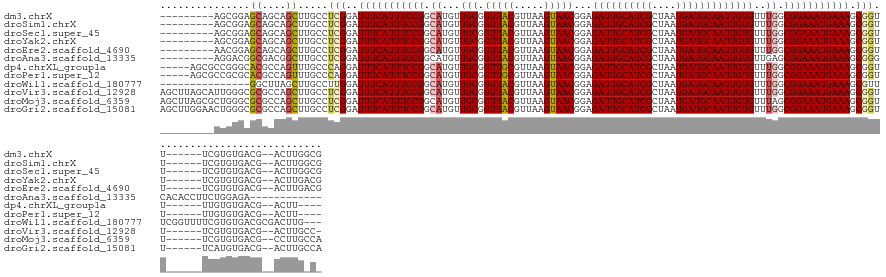
>dm3.chrX 12000568 130 + 22422827 ---------AGCGGAGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGGCG ---------..((..((((....))))..))..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))((.(..------(((.....))--)..).)). ( -46.90, z-score = -1.90, R) >droSim1.chrX 9260480 130 + 17042790 ---------AGCGGAGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGGCG ---------..((..((((....))))..))..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))((.(..------(((.....))--)..).)). ( -46.90, z-score = -1.90, R) >droSec1.super_45 222172 130 - 245362 ---------AGCGGAGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGGCG ---------..((..((((....))))..))..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))((.(..------(((.....))--)..).)). ( -46.90, z-score = -1.90, R) >droYak2.chrX 19920052 130 - 21770863 ---------AGCGGAGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGACG ---------.(((((((.((.....))..((..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))))------))))......--........ ( -45.40, z-score = -1.76, R) >droEre2.scaffold_4690 3919215 130 - 18748788 ---------AACGGAGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGACG ---------.(((((((.((.....))..((..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))))------))))......--........ ( -45.10, z-score = -2.02, R) >droAna3.scaffold_13335 1975023 126 - 3335858 ---------AGGACGGCGACGGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUGAGCGGAAAUGAAAGCGGGCACACCUUCUGGAGA------------ ---------(((.((....))...((((.((..(((((((((((((((....(.(((((.....))))).).((((((((((....))))))))))))))....))))))))))).))))))..)))........------------ ( -45.50, z-score = -2.27, R) >dp4.chrXL_group1a 2720998 130 + 9151740 -----AGCGCCGGGCACGCCAGUUUGCCCAGGAUUUCAUUUCCGGCAUGUUGCGGUUGCGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UUGUGUGACG--ACUU---- -----.((.(((((((........))))).)).(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))))).(((------......))).--....---- ( -47.90, z-score = -1.89, R) >droPer1.super_12 1825301 130 - 2414086 -----AGCGCCGCGCACGCCAGUUUGCCCAGGAUUUCAUUUCCGGCAUGUUGCGGUUGCGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UUGUGUGACG--ACUU---- -----..((.((((((.(((......(....).(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))..))).------.)))))).))--....---- ( -46.10, z-score = -1.22, R) >droWil1.scaffold_180777 3756195 129 + 4753960 ---------------GGCUUAGCUUGCCUUGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGUUUCGGUUUUCGUGUGACGCGACUUG--- ---------------(((.......))).....(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))(((((.((.....))...)))))......--- ( -46.00, z-score = -2.90, R) >droVir3.scaffold_12928 3506667 138 + 7717345 AGCUUAGCAUUGGGCGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCGUGUGACG--ACUUGCC- ......((.((((((((....))..))).))).(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))))(((.------(((.....))--)...)))- ( -47.10, z-score = -0.94, R) >droMoj3.scaffold_6359 53265 139 + 4525533 AGCUUAGCGCUGGGCGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUAGCGGAAAUGAAAGCGGUU------UCGUGUGACG--CCUUGCCA ......(.((.((((((((..((..(((.....(((((((((((((((....(.(((((.....))))).).((((((((((....))))))))))))))....)))))))))))..))).------..))))).))--))).))). ( -49.50, z-score = -0.81, R) >droGri2.scaffold_15081 53143 139 - 4274704 AGCUUGGAACUGGGCGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU------UCAUGUGACG--ACUUGCCA .((.(((((((((((((....))..))).....(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))))------))).))....--........ ( -50.20, z-score = -1.82, R) >consensus _________AGGGGAGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUU______UCGUGUGACG__ACUUG_C_ ...............((....)).....(((..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))............................ (-35.17 = -35.15 + -0.02)
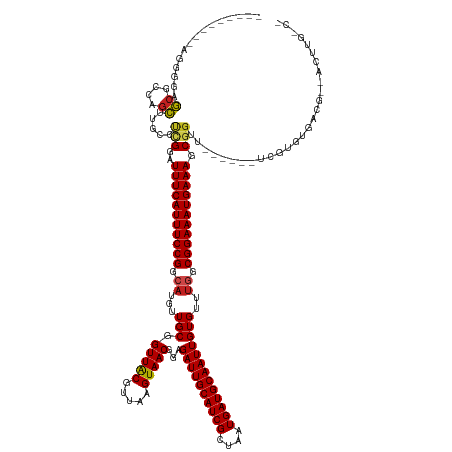
| Location | 12,000,568 – 12,000,698 |
|---|---|
| Length | 130 |
| Sequences | 12 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 86.05 |
| Shannon entropy | 0.29754 |
| G+C content | 0.49643 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -27.87 |
| Energy contribution | -27.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12000568 130 - 22422827 CGCCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCUCCGCU--------- .(((...(--((.....)))------...((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)).))).(((....))).....--------- ( -38.10, z-score = -3.30, R) >droSim1.chrX 9260480 130 - 17042790 CGCCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCUCCGCU--------- .(((...(--((.....)))------...((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)).))).(((....))).....--------- ( -38.10, z-score = -3.30, R) >droSec1.super_45 222172 130 + 245362 CGCCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCUCCGCU--------- .(((...(--((.....)))------...((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)).))).(((....))).....--------- ( -38.10, z-score = -3.30, R) >droYak2.chrX 19920052 130 + 21770863 CGUCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCUCCGCU--------- .((....(--((.....)))------....))(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..((..(((......)))..))..--------- ( -35.60, z-score = -2.56, R) >droEre2.scaffold_4690 3919215 130 + 18748788 CGUCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCUCCGUU--------- .((....(--((.....)))------....))(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..((..(((......)))..))..--------- ( -35.80, z-score = -2.70, R) >droAna3.scaffold_13335 1975023 126 + 3335858 ------------UCUCCAGAAGGUGUGCCCGCUUUCAUUUCCGCUCAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCCGUCGCCGUCCU--------- ------------......((.(((.((((((.(((((((((((.........((((((((......))))))))....(((((.....))))).((.....)))))))))))))..)).)))).))).))........--------- ( -41.60, z-score = -4.36, R) >dp4.chrXL_group1a 2720998 130 - 9151740 ----AAGU--CGUCACACAA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGCAACCGCAACAUGCCGGAAAUGAAAUCCUGGGCAAACUGGCGUGCCCGGCGCU----- ----.((.--((((......------......(((((((((((.(((((...((((((((......))))))))....))).......((....))....)).)))))))))))....(((((........)))))))))))----- ( -39.70, z-score = -2.88, R) >droPer1.super_12 1825301 130 + 2414086 ----AAGU--CGUCACACAA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGCAACCGCAACAUGCCGGAAAUGAAAUCCUGGGCAAACUGGCGUGCGCGGCGCU----- ----..((--((((((....------..(((.(((((((((((.(((((...((((((((......))))))))....))).......((....))....)).)))))))))))...)))((......))))).)))))...----- ( -39.40, z-score = -2.26, R) >droWil1.scaffold_180777 3756195 129 - 4753960 ---CAAGUCGCGUCACACGAAAACCGAAACGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCAAGGCAAGCUAAGCC--------------- ---...(((((((....((.....))..))))(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).))))))))))).....)))..........--------------- ( -35.10, z-score = -3.81, R) >droVir3.scaffold_12928 3506667 138 - 7717345 -GGCAAGU--CGUCACACGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCGCCCAAUGCUAAGCU -.((...(--((.....)))------...((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..))..)).((((((((.......)))).)))) ( -38.20, z-score = -1.65, R) >droMoj3.scaffold_6359 53265 139 - 4525533 UGGCAAGG--CGUCACACGA------AACCGCUUUCAUUUCCGCUAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCGCCCAGCGCUAAGCU .......(--(.((...((.------...)).(((((((((((.........((((((((......))))))))....(((((.....))))).((.....)))))))))))))...)).)).(((((((((.....))))).)))) ( -44.10, z-score = -2.19, R) >droGri2.scaffold_15081 53143 139 + 4274704 UGGCAAGU--CGUCACAUGA------AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCGCCCAGUUCCAAGCU ((((....--.)))).....------....(((((((((((((.((......((((((((......))))))))....(((((.....))))).......)).))))))))).......((..((((((.....)))))))))))). ( -39.10, z-score = -1.80, R) >consensus _G_CAAGU__CGUCACACGA______AACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCCGCGCCCCU_________ ..............................(((((((((((((.((......((((((((......))))))))....(((((.....))))).......)).))))))))))......)))..((....))............... (-27.87 = -27.79 + -0.08)

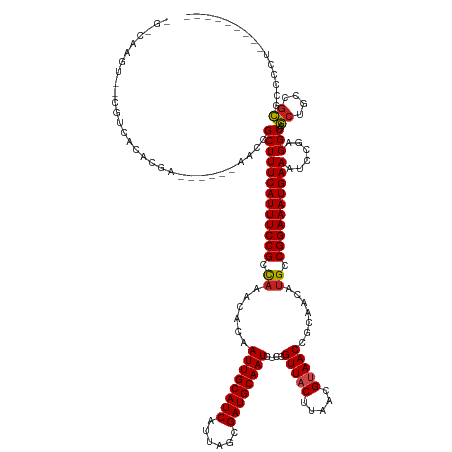
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:11 2011