| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,118,974 – 11,119,091 |
| Length | 117 |
| Max. P | 0.931013 |

| Location | 11,118,974 – 11,119,091 |
|---|---|
| Length | 117 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Shannon entropy | 0.62067 |
| G+C content | 0.48402 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.90 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
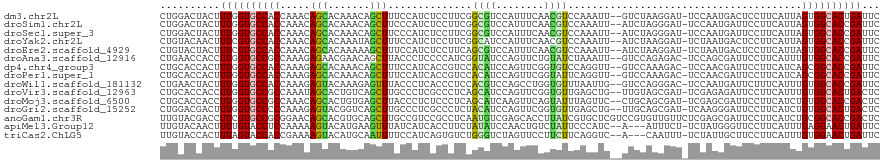
>dm3.chr2L 11118974 117 + 23011544 CUGGACUACUUUGGUGCCACCAAACAGCACAAACAGCUUUCCAUCUCCUUCGGCGUCCAUUUCAACGUCCAAAUU--GUCUAAGGAU-UCCAAUGACUCCUUCAUUAGUGGCACUGAUUC ..........(..((((((((((..(((.......))).............(((((........)))))....))--)...(((((.-((....)).))))).....)))))))..)... ( -25.10, z-score = -1.10, R) >droSim1.chr2L 10918571 117 + 22036055 CUGGACUACUUUGGUGCCACCAAACAGCACAAACAGCUUCCCAUCUCCUUCGGCGUCCAUUUCAACGUCCAAAUU--AUCUAGGGAU-UCCAAUGAUUCCUUCAUUAGUGGCACCGAUUC ..........((((((((((.....(((.......)))((((.........(((((........)))))......--.....)))).-...(((((.....))))).))))))))))... ( -29.35, z-score = -2.54, R) >droSec1.super_3 6520366 117 + 7220098 CUGGACUACUUUGGUGCCACCAAACAGCACAAACAGCUUCCCAUCUCCUUCGGCGUCCAUUUCAACGUCCAAAUU--AUCUAGGGAU-UCCAAUGAUUCCUUCAUUAGUGGCACCGAUUC ..........((((((((((.....(((.......)))((((.........(((((........)))))......--.....)))).-...(((((.....))))).))))))))))... ( -29.35, z-score = -2.54, R) >droYak2.chr2L 7515131 117 + 22324452 CUGUACAACUUUCGUGCCACCAAACAGCACAAAUAGCUUUCCAUCUCCUUCGGCAUCCAUUUCAACGUCCAAAUU--AUCUAAGGAU-UCUAAUGACUCCUUCAUUAGUGGCACCGAUUC ...........(((((((((.....(((.......))).....................................--....(((((.-((....)).))))).....)))))).)))... ( -19.60, z-score = -1.69, R) >droEre2.scaffold_4929 12315838 117 - 26641161 CUGUACUACUUUCGUGCCACCAAACAGCACAAAAAGCUUUCCAUCUCCUUCAGCGUCCAUUUCAACGUCCAAAUU--AUCUAAGGAU-UCUAAUGACUCCUUCAUUAGUGGCACCGAUUC ...........(((((((((.....(((.......)))..............((((........)))).......--....(((((.-((....)).))))).....)))))).)))... ( -20.80, z-score = -2.36, R) >droAna3.scaffold_12916 12243630 117 + 16180835 CUGAACCACCUUGGUGCCGCCAAAGAGAACGAACAGCUUACCCUCCCCAUCGGUAUCCAGUUCUGUAUCUAAAUU--GUCCAGAGAC-UCCAGCGAUUCCUUCAUUUGUGGCACCGAUUC ..........((((((((((.(((..(((.(((..(((((((.........))))...(((((((...(......--)..)))).))-)..)))..))).))).)))))))))))))... ( -29.40, z-score = -1.87, R) >dp4.chr4_group3 6470398 117 + 11692001 CUGCACCACUUUGGUGCCACCAAAGAGCACAAACAGCUUUCCAUCACCGUCCACAUCCAGUUCGGUGUCCAGGUU--GUCCAAAGAC-UCCAACGAUUCCUUCAUCAGCGGCACCGAUUC ..........((((((((......((((....(((((((.....(((((..(.......)..)))))...)))))--)).....).)-))....(((......)))...))))))))... ( -29.50, z-score = -2.05, R) >droPer1.super_1 7961939 117 + 10282868 CUGCACCACUUUGGUGCCACCAAAGAGCACAAACAGCUUUCCAUCACCGUCCACAUCCAGUUCGGUAUUCAGGUU--GUCCAAAGAC-UCCAACGAUUCCUUCAUCAGCGGCACCGAUUC ..........((((((((.....(((((.......)))))..(((((((..(.......)..)))).....((..--(((....)))-.))...)))............))))))))... ( -27.10, z-score = -2.02, R) >droWil1.scaffold_181132 429463 117 - 1035393 CUGAACUACUUUGGUGCCACCAAAGAGUACAAAGAGUUUACCCUCACCCUCCACGUCCAGCCUGGUGUUUAAUUG--GUCCAGGGAC-UCCAAUGAUUCUUUCAUUUGUGGCACCGAUUC ..........((((((((((.(((((((.....((((...((((.(((....(((.((.....)))))......)--))..))))))-)).....))))))).....))))))))))... ( -33.90, z-score = -1.86, R) >droVir3.scaffold_12963 12299490 117 - 20206255 CUGCACCACCUUGGUGCCGCCAAAUAGCACUGUCAGCUUGCCCUCGCCCUCAGCAUCCAGUUCGGUGUUGAGCUG--UUGUAGCGAU-UCGAGAGAUUCCUUCAUUUGUGGCACUGACUC ..(((((.....))))).((......))...(((((..((((......(((((((((......)))))))))...--.((.((.((.-((....)).)))).)).....))))))))).. ( -36.70, z-score = -1.14, R) >droMoj3.scaffold_6500 2947382 117 + 32352404 CUGCACCACCUUGGUGCCGCCAAACAGCACUGUGAGCUUACCCUCUCCCUCAGCAUCAAGUUCAGUAUUUAGUUC--CUGCAGCGAU-UCGAGCGAUUCCUUCAUCUGUGGCACCGACUC ..........((((((((((....(((.(((((((((((..................))))))).....))))..--)))..((...-....)).............))))))))))... ( -25.27, z-score = 0.36, R) >droGri2.scaffold_15252 6211881 117 - 17193109 CUGGACGACUUUGGUGCCCCCAAAGAGUACGGUCAGCUUGCCCUCGCCCUCUACAUCCAGUUCGGUGUUGAGCUG--UUGCAGCGAU-UCAAGGGAUUCCUUCAUCUGUGGCACUGACUC (((((...((((((.....)))))).(((.(((.((......)).)))...))).))))).((((((((..((((--...))))(((-..((((....)))).)))...))))))))... ( -32.50, z-score = 0.89, R) >anoGam1.chr3R 47980216 120 - 53272125 UUGUACGACCUUCGUGCCGCGGAACAGCACGUGCAGCUUGCCGUCCGCCUCAAUGUCGAGCACCUUAUCGUGCUCGUCCGUGUUGUUCUCGAGCGAUUCCUUCAUCUGCGGCACCGACUC ...........((((((((((((((((((((..(.....((.....)).........((((((......)))))))..))))))))))).((..((.....)).)).)))))).)))... ( -43.40, z-score = -2.35, R) >apiMel3.Group12 6924824 114 - 9182753 UUGUACAACUUUUGUACCUCCAAAAAGUACAUGAAGUUUAUCAUCACCUUCUAUAUCCAACUGUCUAUUCCCAUC--A---AUUUCU-UCUAUGGGUUCCUUCAUUUGAGGAACUGAUUC .((...(((((((((((.........))))).))))))...))................................--.---......-...((.((((((((.....)))))))).)).. ( -17.00, z-score = -0.38, R) >triCas2.ChLG5 114239 114 - 18847211 UUGUACCACUUUAGUACCACCGAAAAGUACAUGCAAUUUUCCAUCAGUGUCUGGGUCUAGUUCCUUCUUCAGGUC--A---CAAUUU-UCUAUUGCUUCCUUCAUUUGUGGAACUGAUUC ..........(((((.((((.((.(((.....(((((.........(((((((((............)))))).)--)---).....-...)))))...)))..)).)))).)))))... ( -18.63, z-score = 0.40, R) >consensus CUGUACCACUUUGGUGCCACCAAACAGCACAAACAGCUUUCCAUCUCCUUCGGCAUCCAGUUCAGUGUCCAAAUU__AUCUAGGGAU_UCCAAUGAUUCCUUCAUUUGUGGCACCGAUUC ..........((((((((((.....(((.......)))..............((((........)))).......................................))))))))))... (-13.95 = -13.05 + -0.90)
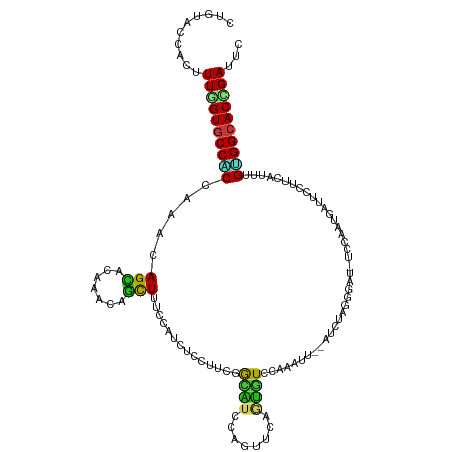
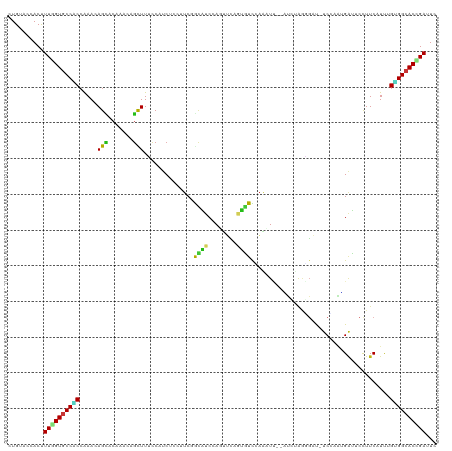
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:36 2011