
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,989,865 – 11,989,958 |
| Length | 93 |
| Max. P | 0.824225 |

| Location | 11,989,865 – 11,989,958 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.51789 |
| G+C content | 0.48784 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824225 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11989865 93 + 22422827 CUUUUGCUUUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCGUCGUCGCUGUCGAUGGAGAGAUGGUGCC---- .....((.((((((((((....)))))).((((....))))...))))((((((..(((.((.((....)).)).)))..).)))))...)).---- ( -28.50, z-score = -1.78, R) >droSim1.chrX 9249977 86 + 17042790 CUUUUGCUUUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCGU---CGCUGUCGAUGGA----UGGUGCC---- .....(((((((((((((....)))))).((((....))))...))))).......(((((..(((---((....)))))))----))).)).---- ( -22.40, z-score = -0.96, R) >droSec1.super_45 211650 86 - 245362 CUUUUGCUUUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCGU---CGCUGUCGAUGGAA----GGUGCC---- .....(((((.(((((((....))))))..(((((..(((..(((.....(....)....)))..)---))..))))).).))----)))...---- ( -24.60, z-score = -1.68, R) >droYak2.chrX 19909286 79 - 21770863 CUUUUGCUUUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCGC------UGUCGAUGGUGCC------------ ..........((((((((....))))))..(((((..(((..((((((.....))).)))..))).------.)))))....)).------------ ( -21.90, z-score = -2.08, R) >droEre2.scaffold_4690 3908790 79 - 18748788 CUUUUGCUCUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGCCGC------UGUCGAUGGUGCC------------ ..(((((.....((((((....)))))).((((....))))...))))).....(..((((((...------.).)))))..)..------------ ( -19.30, z-score = -0.75, R) >dp4.chrXL_group1a 2709125 76 + 9151740 CUUUUGCUUCGCGGUGGCUUAUUCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCCUCGUCCGCGUC--------------------- .........(((((((((.......))))..((((...((...........))..))))..........)))))..--------------------- ( -14.50, z-score = -0.77, R) >droPer1.super_12 1813174 76 - 2414086 CUUUUGCUUCGCGGUGGCUUAUUCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCCUCGUCCGCGUC--------------------- .........(((((((((.......))))..((((...((...........))..))))..........)))))..--------------------- ( -14.50, z-score = -0.77, R) >droVir3.scaffold_12928 3494685 91 + 7717345 CUUUUGCUUUGCGGUGGCUUAUGUCGCCGUUCGAUUUUGAAAAUGUAAAUCUCUUGU--CC-UCAUCCAUGUGACCC---GCCCCCUACCCGGAAGC .....(((((((((((((....))))))))..(((((.........)))))....((--((-........).)))..---............))))) ( -18.10, z-score = -0.56, R) >droGri2.scaffold_15081 40639 95 - 4274704 CUUUUGCUUUGCGGUGGCUUAUGUCGCCAUUCGAUUUUCAAAAUGUAAAUCUCUUGU--UCACCAUUCACGUGCCCCACAGCCACCCACUCAAUAAC ...(((...((.(((((((..((..(((....(((((.(.....).)))))...((.--....)).....).))..)).)))))))))..))).... ( -16.10, z-score = -0.91, R) >consensus CUUUUGCUUUGCGGUGGCUUAUGCCGCCAUUCGAUUUUGAAAAUGUAAAUCUCUUGUCGUCGUCGU___CGCUGUCGAUGGA_______________ ..(((((.....((((((....)))))).((((....))))...)))))................................................ (-12.11 = -12.27 + 0.16)

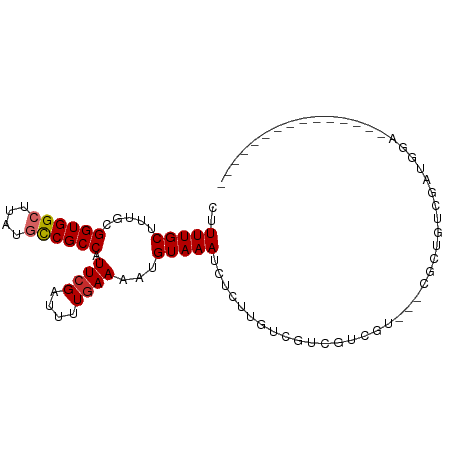
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:08 2011