| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,968,750 – 11,968,849 |
| Length | 99 |
| Max. P | 0.996402 |

| Location | 11,968,750 – 11,968,849 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.70 |
| Shannon entropy | 0.65762 |
| G+C content | 0.44748 |
| Mean single sequence MFE | -12.74 |
| Consensus MFE | -8.37 |
| Energy contribution | -8.59 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
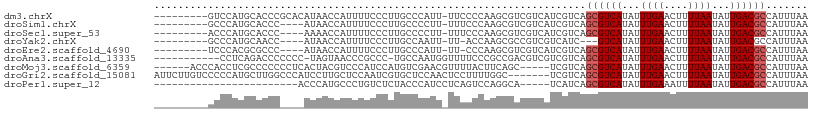
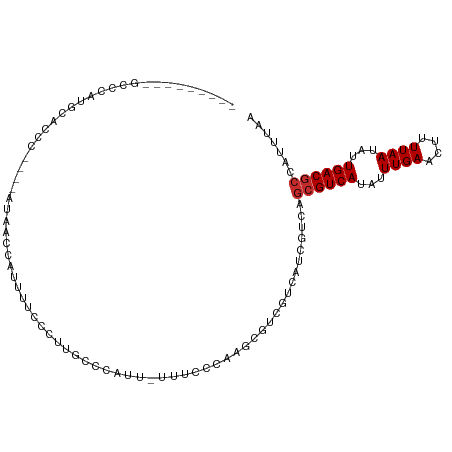
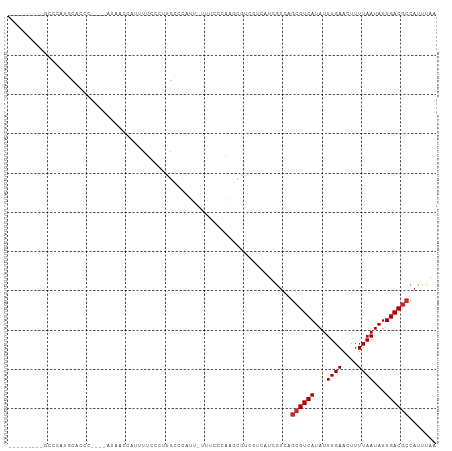
>dm3.chrX 11968750 99 + 22422827 ---------GUCCAUGCACCCGCACAUAACCAUUUUCCCUUGCCCAUU-UUCCCCAAGCGUCGUCAUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ---------(.(.(((.((.(((.........................-........)))..))))).).).((((((...((((....))))...))))))....... ( -12.31, z-score = -1.43, R) >droSim1.chrX 9230806 95 + 17042790 ---------GCCCAUGCACCC----AUAACCAUUUUCCCUUGCCCCUU-UUUCCCAAGCGUCGUCAUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ---------((....))....----.......................-..........(.((....)).).((((((...((((....))))...))))))....... ( -10.70, z-score = -1.39, R) >droSec1.super_53 185109 95 + 190754 ---------ACCCAUGCACCC----AAAACCAUUUUCCCUUGCCCCUU-UUUCCCAAGCGUCGUCAUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ---------............----.......................-..........(.((....)).).((((((...((((....))))...))))))....... ( -10.40, z-score = -1.46, R) >droYak2.chrX 19888554 91 - 21770863 ---------GCCCAUGCAACC----AUAACCAUUUUCCCUUGCCAAUU-UU-ACCAAGCGCCGUCGUCAUC---GUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ---------......((....----.............((((......-..-..)))).............---((((...((((....))))...))))))....... ( -6.20, z-score = 0.80, R) >droEre2.scaffold_4690 3889094 94 - 18748788 ---------UCCCACGCGCCC----AUAACCAUUUUCCCUUGCCCAUU-UU-CCCAAGCGUCGUCAUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ---------....((((((..----.......................-..-.....))).)))........((((((...((((....))))...))))))....... ( -12.55, z-score = -1.95, R) >droAna3.scaffold_13335 1938916 96 - 3335858 -----------CCUCAGACCCCCCC-UAGUAACCCGCCC-UGCCAAUGGUUUUCCCGCCGACGUCGUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA -----------..............-.............-....((((((......((.((((....)))).))((((...((((....))))...))))))))))... ( -17.60, z-score = -1.85, R) >droMoj3.scaffold_6359 16004 98 + 4525533 ------ACCCACCUCGCCCCCCCUCACUACGUCCCAUCCAUGUCGAACGUUUUACUUCAGC-----UCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ------..................................((.(((.(...........).-----))).))((((((...((((....))))...))))))....... ( -12.10, z-score = -1.79, R) >droGri2.scaffold_15081 12385 102 - 4274704 AUUCUUGUCCCCCAUGCUUGGCCCAUCCUUGCUCCAAUCGUGCUCCAACUCCUUUUGGC-------UCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ............((((.((((..((....))..)))).))))..((((......)))).-------......((((((...((((....))))...))))))....... ( -18.40, z-score = -1.96, R) >droPer1.super_12 1794034 80 - 2414086 ------------------------ACCCAUGCCCUGUCUCUACCCAUCCUCAGUCCAGGCA-----UCAUCAGCGUCAUAUUUGAAAUUUUAAUAUUGACGCCAUUUAA ------------------------....((((((((..............)))....))))-----).....((((((...((((....))))...))))))....... ( -14.44, z-score = -2.80, R) >consensus _________GCCCAUGCACCC____AUAACCAUUUUCCCUUGCCCAUU_UUUCCCAAGCGUCGUCAUCGUCAGCGUCAUAUUUGAACUUUUAAUAUUGACGCCAUUUAA ........................................................................((((((...((((....))))...))))))....... ( -8.37 = -8.59 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:04 2011