| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,964,812 – 11,964,907 |
| Length | 95 |
| Max. P | 0.958306 |

| Location | 11,964,812 – 11,964,907 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.65380 |
| G+C content | 0.51387 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -12.11 |
| Energy contribution | -11.45 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11964812 95 + 22422827 AACGUUGGCGACGUCCUGUACUUUGGGGUACAAUUAGGCAUCGAACACGAUGGGGAGAUACCGAAUACGUUCGGAAUAUUCACCGUUAAAUCAUC------------------- ..(((((.(((.(((.((((((....))))))....))).)))....)))))((..(((((((((....)))))..))))..))...........------------------- ( -24.50, z-score = -0.52, R) >droSim1.chrX 9228224 95 + 17042790 GUGGCGGGCGACAUCCUGUACUUUGGGGUACAGUUAAGCAUCGAUCACGAUGGAGAGGUCCCGAAUACGUUCGGAAUAUUCACCGUUAAAUCAUU------------------- .((((((..(((.(((((((((....))))))).....(((((....)))))..)).)))(((((....)))))........)))))).......------------------- ( -29.10, z-score = -1.44, R) >droEre2.scaffold_4690 3884951 108 - 18748788 UCCGUGGGCGAUGAUCUGUGCUUUGGGGUUAAACUAUACGGCGAGGUCGCUGAGGAGAUCCCGAUUACGUUCGGGAUUUUCACCGUUAAGGAGGAGGAGCUGCACCAC------ ...(((((((.(..(((...(((((((((.........(((((....))))).(((((((((((......)))))))))))))).)))))).)))..)..))).))))------ ( -41.30, z-score = -2.26, R) >droYak2.chrX 19883914 114 - 21770863 UCCGUGGGCGAUGAUCUCUGUUUUGGGGUCAAAAUAAACGGCGAGGUCGCCGUGGAGAUACCGGUUACGUUCGGGAUCUUCACCGUCAAGGCGGAGGAGCUGCAUUCCCAUCAC ...(((((.((((.(((((((((((((((........((((((....))))))((((((.((((......)))).))))))))).)))))))))))).....)))))))))... ( -47.90, z-score = -2.92, R) >droSec1.super_53 181378 95 + 190754 GUGGCGGGCGACAUCCUGUACUUUGGGGUACAAUUAAGCAUCGAUCACGAUGGGGAGAUCCCGAAUACGUUCGGAAUAUUCACCGUUAAAUCAUA------------------- .((((((..((..(((((((((....))))).......(((((....)))))))))..))(((((....)))))........)))))).......------------------- ( -28.30, z-score = -1.27, R) >droAna3.scaffold_13335 1933349 101 - 3335858 UUCGUUGGCGAUGACAUAUCCUUUGGCGUGGAUAUUGAUCCGUCGGUGGAGCAACAGCUACCAUGGACCUUCGGUAUCUUCACCGUCAAGGCGGAGCCGGC------------- ...((((((((((...))))((((((((.(((((((((...(((.(((((((....))).)))).)))..)))))))))....))))))))....))))))------------- ( -40.30, z-score = -2.42, R) >consensus UUCGUGGGCGACGACCUGUACUUUGGGGUACAAUUAAACAUCGAGCACGAUGGGGAGAUACCGAAUACGUUCGGAAUAUUCACCGUUAAAGCAGA___________________ ......((((............................(((((....)))))........((((......)))).........))))........................... (-12.11 = -11.45 + -0.66)

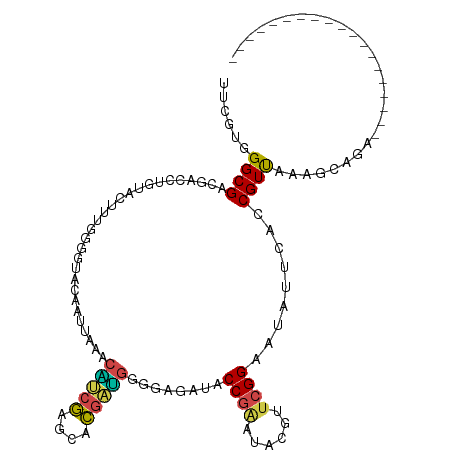
| Location | 11,964,812 – 11,964,907 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.65380 |
| G+C content | 0.51387 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
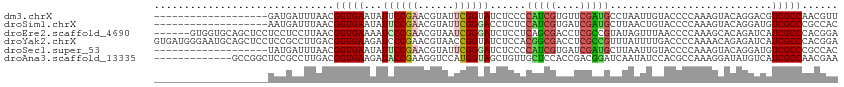
>dm3.chrX 11964812 95 - 22422827 -------------------GAUGAUUUAACGGUGAAUAUUCCGAACGUAUUCGGUAUCUCCCCAUCGUGUUCGAUGCCUAAUUGUACCCCAAAGUACAGGACGUCGCCAACGUU -------------------...........(((((.....(((((....)))))....(((.(((((....)))))......(((((......))))))))..)))))...... ( -22.40, z-score = -0.79, R) >droSim1.chrX 9228224 95 - 17042790 -------------------AAUGAUUUAACGGUGAAUAUUCCGAACGUAUUCGGGACCUCUCCAUCGUGAUCGAUGCUUAACUGUACCCCAAAGUACAGGAUGUCGCCCGCCAC -------------------...........((((......(((((....)))))(((.((..(((((....))))).....((((((......)))))))).)))...)))).. ( -27.00, z-score = -2.05, R) >droEre2.scaffold_4690 3884951 108 + 18748788 ------GUGGUGCAGCUCCUCCUCCUUAACGGUGAAAAUCCCGAACGUAAUCGGGAUCUCCUCAGCGACCUCGCCGUAUAGUUUAACCCCAAAGCACAGAUCAUCGCCCACGGA ------((((.((...............(((((((..(((((((......))))))).((......))..)))))))...((((.......))))..........))))))... ( -28.30, z-score = -1.14, R) >droYak2.chrX 19883914 114 + 21770863 GUGAUGGGAAUGCAGCUCCUCCGCCUUGACGGUGAAGAUCCCGAACGUAACCGGUAUCUCCACGGCGACCUCGCCGUUUAUUUUGACCCCAAAACAGAGAUCAUCGCCCACGGA ....(((((......((.(.(((......))).).)).)))))..(((...(((((((((.((((((....))))))...(((((....)))))..))))).))))...))).. ( -31.40, z-score = -0.20, R) >droSec1.super_53 181378 95 - 190754 -------------------UAUGAUUUAACGGUGAAUAUUCCGAACGUAUUCGGGAUCUCCCCAUCGUGAUCGAUGCUUAAUUGUACCCCAAAGUACAGGAUGUCGCCCGCCAC -------------------...........((((......(((((....)))))(((.(((.(((((....)))))......(((((......)))))))).)))...)))).. ( -25.30, z-score = -1.39, R) >droAna3.scaffold_13335 1933349 101 + 3335858 -------------GCCGGCUCCGCCUUGACGGUGAAGAUACCGAAGGUCCAUGGUAGCUGUUGCUCCACCGACGGAUCAAUAUCCACGCCAAAGGAUAUGUCAUCGCCAACGAA -------------...(((...)))(((.((((((.(((.(((..(((....((.(((....))))))))..)))))).((((((........)))))).)))))).))).... ( -31.60, z-score = -1.08, R) >consensus ___________________UACGACUUAACGGUGAAUAUUCCGAACGUAUUCGGGAUCUCCCCAUCGACCUCGAUGCUUAAUUGUACCCCAAAGUACAGGACAUCGCCCACGAA ..............................(((((...((((((......))))))......(((((....)))))...........................)))))...... (-15.36 = -15.42 + 0.06)
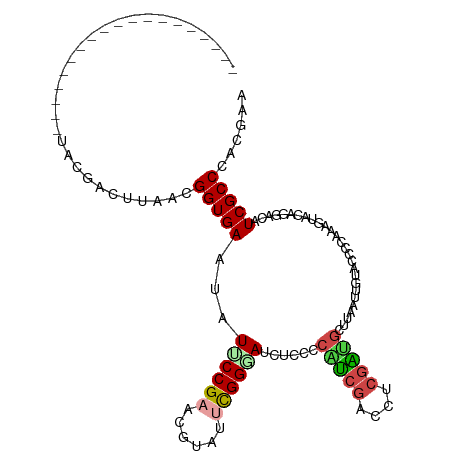
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:36:04 2011