| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,935,258 – 11,935,385 |
| Length | 127 |
| Max. P | 0.948268 |

| Location | 11,935,258 – 11,935,385 |
|---|---|
| Length | 127 |
| Sequences | 11 |
| Columns | 145 |
| Reading direction | forward |
| Mean pairwise identity | 84.70 |
| Shannon entropy | 0.31272 |
| G+C content | 0.44730 |
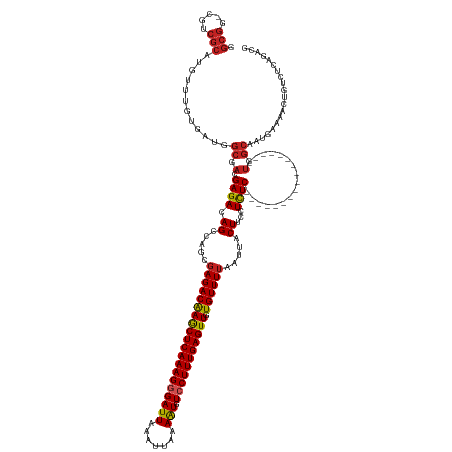
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
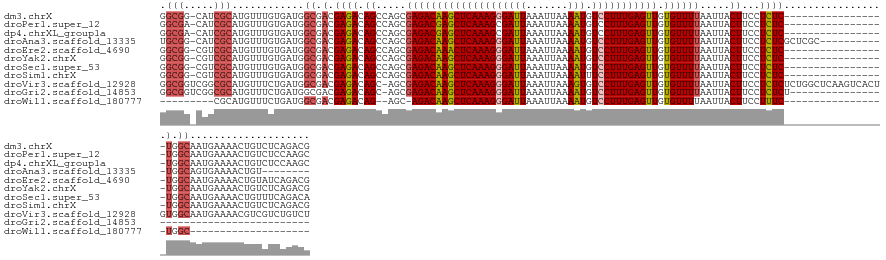
>dm3.chrX 11935258 127 + 22422827 GGCGG-CAUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUCUCAGACG ..((.-(((((((....))))))).))..(((((((((((.(((...((((((((((((((.......))).)))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -46.20, z-score = -4.08, R) >droPer1.super_12 1754904 127 - 2414086 GGCGA-CAUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACGAGCUCAAAGCGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUCUCCAAGC ..((.-(((((((....))))))).))..(((((((((((.(((...(((((((((.(((...........))))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -41.50, z-score = -3.02, R) >dp4.chrXL_group1a 2655059 127 + 9151740 GGCGA-CAUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACGAGCUCAAAGCGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUCUCCAAGC ..((.-(((((((....))))))).))..(((((((((((.(((...(((((((((.(((...........))))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -41.50, z-score = -3.02, R) >droAna3.scaffold_13335 1902027 125 - 3335858 UGCGG-CAUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUCGCUCGC-----------UGGCAGUGAAAACUGU-------- ..((.-(((((((....))))))).))......((((.(((((((..((((((((((((((.......))).)))))))))))(((.......)))....))))))).))-----------))(((((....)))))-------- ( -50.20, z-score = -4.82, R) >droEre2.scaffold_4690 3860603 127 - 18748788 GGCGG-CGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAACUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUAUCAGACG ..((.-(((((((....))))))).))..((.((((((((.(((...((((((((((((((.......))).)))))))))))(((.......)))...))).)-----------------))))..(....).))).))..... ( -39.60, z-score = -2.51, R) >droYak2.chrX 19852754 127 - 21770863 GGCGG-CGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUCUCAGACG ..((.-(((((((....))))))).))..(((((((((((.(((...((((((((((((((.......))).)))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -45.80, z-score = -3.75, R) >droSec1.super_53 153094 127 + 190754 GGCGG-CGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUUUCAGACA ..((.-(((((((....))))))).))..(((((((((((.(((...((((((((((((((.......))).)))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -43.20, z-score = -3.06, R) >droSim1.chrX 9197697 127 + 17042790 GGCGG-CGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUUUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC-----------------UGGCAAUGAAAACUGUCUCAGACG ..((.-(((((((....))))))).))..(((((((((((.(((...((((((((((((............))))))))))))(((.......)))...))).)-----------------))))..(....).))))))..... ( -47.40, z-score = -4.40, R) >droVir3.scaffold_12928 3419725 144 + 7717345 GGCGGUCGGCGCAUGUUUCUGAUGGCGACGAGACAGC-AGCGAGACAAGCUCAAAGGGAUUAAAUUAAAGUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUCUCUGGCUCAAGUCACUGUGGCAAUGAAAACGUCGUCUGUCU (((((.(((((....((((.....((.(((.((((((-((.((((..((((((((((((((.......))).)))))))))))(((.......)))....)))).)).)))...)))..))).))...)))).))))).))))). ( -44.80, z-score = -1.22, R) >droGri2.scaffold_14853 10126694 104 - 10151454 GGCGGUCGGCGCAUGUUUCUGAUGGCGACGAGACAGC-AGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUCU---------------------------------------- ((.(((..((((.((((((..........))))))))-.))((((((((((((((((((((.......))).))))))))))).))))))....))).)).....---------------------------------------- ( -29.40, z-score = -0.97, R) >droWil1.scaffold_180777 1078009 96 - 4753960 ---------CGCAUGUUUCUGAUGGCGACGAGACAG--AGC-AGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUUUC-----------------UGGC-------------------- ---------.((.((((((..........)))))).--.))-(((((((((((((((((((.......))).))))))))))).))))).........((....-----------------.)).-------------------- ( -23.00, z-score = -0.73, R) >consensus GGCGG_CGUCGCAUGUUUGUGAUGGCGACGAGACAGCCAGCGAGACAAGCUCAAAGGGAUUAAAUUAAAAUGUCCUUUGAGUUGUGUUUUAAUUACUUCCUCUC_________________UGGCAAUGAAAACUGUCUCAGACG .(((.....))).((((......))))..(((..((.....((((((((((((((((((((.......))).))))))))))).)))))).....))..)))........................................... (-21.60 = -21.75 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:56 2011