
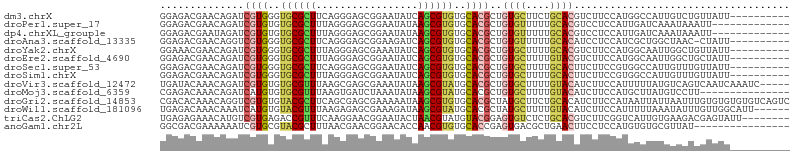
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,915,181 – 11,915,276 |
| Length | 95 |
| Max. P | 0.748346 |

| Location | 11,915,181 – 11,915,276 |
|---|---|
| Length | 95 |
| Sequences | 14 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.63135 |
| G+C content | 0.47574 |
| Mean single sequence MFE | -29.06 |
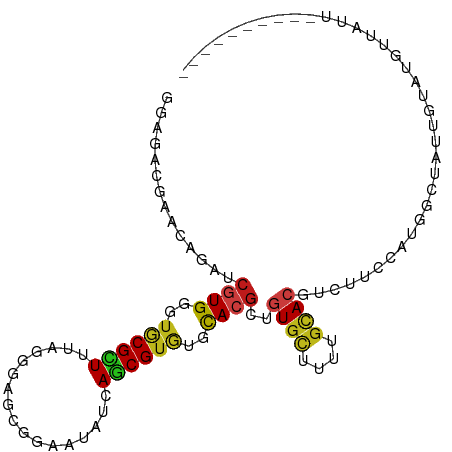
| Consensus MFE | -13.28 |
| Energy contribution | -13.01 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11915181 95 - 22422827 GGAGACGAACAGAUCGUGGGUGCGCUUCAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUCUGCACGUCUUCCAUGGCCAUUGUCUGUUAUU---------- (....).(((((((.(((.((((((((....))))((((...((((((....))))))...))))))))(((......)))))).)))))))...---------- ( -36.80, z-score = -1.75, R) >droPer1.super_17 53411 92 - 1930428 GGAGACGAACAGAUCGUGUGUGCGCUUUAGGGAGCGGAAUAUAAGCGUGUGCACGCUGUGUUUUUGCACGUCCUCCAUUGAUCAAAUAAAUU------------- (((((((.((((..((((((..(((((...............)))))..))))))))))((....)).))).))))................------------- ( -28.26, z-score = -1.42, R) >dp4.chrXL_group1e 3118958 92 - 12523060 GGAGACGAAUAGAUCGUGUGUGCGCUUUAGGGAGCGGAAUAUAAGCGUGUGCACGCUGUGUUUUUGCACGUCCUCCAUUGAUCAAAUAAAUU------------- ((((((((.....))))..((((((((....))))((((((((.((((....)))))))))))).))))...))))................------------- ( -28.20, z-score = -1.62, R) >droAna3.scaffold_13335 1874935 94 + 3335858 GGAGACGAACAGGUCGUGGGUGCGCUUCAGGGAGCGGAAGAUCAGCGUGUGCACGCUGUGUUUUUGCACAUCCUCCAUCGCUGGCUAAC-CUAUU---------- ((((((((.....))))..((((((((....)))).((((((((((((....)))))).))))))))))...)))).............-.....---------- ( -31.60, z-score = -0.23, R) >droYak2.chrX 19832877 95 + 21770863 GGAAACGAACAGAUCGUGGGUGCGCUUUAGGGAGCGAAAUAUCAGCGUGUGCACGCUGUGCUUUUGCACGUCUUCCAUGGCAAUUGGCUGUUAUU---------- (....).(((((.((((....))(((...(((((((......((((((....))))))(((....))))).)))))..)))....)))))))...---------- ( -29.30, z-score = -0.37, R) >droEre2.scaffold_4690 3840009 95 + 18748788 GGAGACGAACAGAUCGUGGGUGCGCUUUAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUUUGUACGUCUUCCAUGGCAAUUGGCUGCUAUU---------- (((((((.(((((.((..(((((((......((........)).....)))))).)..))..))))).))))))).((((((......)))))).---------- ( -32.60, z-score = -1.06, R) >droSec1.super_53 133273 95 - 190754 GGAGACGAACAGAUCGUGGGUGCGCUUCAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUUUGCACUUCUUCCGUGGCCAUUGUUUGUUAUU---------- (....).(((((((.((((.(((((((....))))((((....(((((....)))))((((....))))...))))))).)))).)))))))...---------- ( -36.90, z-score = -2.35, R) >droSim1.chrX 9176714 95 - 17042790 GGAGACGAACAGAUCGUGGGUGCGCUUUAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUUUGCACUUCUUCCGUGGCCAUUGUUUGUUAUU---------- (....).(((((((.((((.(((((((....))))((((....(((((....)))))((((....))))...))))))).)))).)))))))...---------- ( -35.80, z-score = -2.32, R) >droVir3.scaffold_12472 202865 100 - 763072 UGAUACAAACAGAUCGUGUGUGCGUUUAAGCGAGCGAAAUAUAAGCGUAUGCACGCUGUGCUUUUGUACAUCUUCCAUUUUUAUGUCAGUCAAUCAAAUC----- ((((((..(((...((((((((((((((.............)))))))))))))).(((((....))))).............)))..))..))))....----- ( -22.92, z-score = -0.44, R) >droMoj3.scaffold_6359 3399594 90 - 4525533 CGAGACAAACAGAUCAUGUGUGCGUUUAAGUGAUCUAAAUAUAAGCGUAUGCACGCUGUGCUUUUGUACAUCUUCCAUGCUUAUGUCCUU--------------- ...((((.........((((((((((((.((.......)).)))))))))))).(.(((((....))))).)...........))))...--------------- ( -20.70, z-score = -0.84, R) >droGri2.scaffold_14853 3335705 105 + 10151454 CGACACAAACAGGUCGUGUGUACGCUUCAGCGAGCGAAAAAUAAGCGUGUGCACGCUAUGCUUCUGCACAUCUUCCAUAAUUAUUAAUUUGUGUGUGUGUCAGUC .((((((.(((((.((((..(((((((...............)))))))..)))))).(((....))).......................))).)))))).... ( -31.16, z-score = -1.40, R) >droWil1.scaffold_181096 9376160 99 - 12416693 UGAGACAAACAAAUCAUGUGUACGUUUAAGAGAGCGAAAGAUAAGCGUAUGCACGCUAUGCUUUUGUACAUCUUCCAUUUUUAAAUAUUUGUUGGCAUU------ ...(...(((((((.((..........((((..(((((((...(((((....)))))...)))))))...))))..........)))))))))..)...------ ( -19.35, z-score = -0.21, R) >triCas2.ChLG2 6207031 97 - 12900155 UGAGAGAAACAUGUCGUGAGACCGUUUCAAGGAACGGAAUACUAACGUAUGUACGGAGUGUCUCUGCACGUCUUCGGUCAUUGUGAAGACGAGUAUU-------- ..(((((..(((..((((...((((((....)))))).((((....)))).))))..))))))))((.((((((((.......)))))))).))...-------- ( -32.60, z-score = -2.33, R) >anoGam1.chr2L 31347250 89 + 48795086 GGCGACGAAAAAAUCGUGCGUACGCUUUAACGAACGGAACACCAACGUGUGCACCGAGUGACGCUGAACUUCCUCCAUGUGUGCGUUAU---------------- .(((((((.....)))).))).............(((.((((....))))...))).(((((((...((.........))..)))))))---------------- ( -20.70, z-score = 0.89, R) >consensus GGAGACGAACAGAUCGUGGGUGCGCUUUAGGGAGCGGAAUAUCAGCGUGUGCACGCUGUGCUUUUGCACGUCUUCCAUGGCUAUUGUAUGUUAUU__________ ..............((((..((((((.................))))))..))))..((((....)))).................................... (-13.28 = -13.01 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:54 2011