| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,104,838 – 11,104,929 |
| Length | 91 |
| Max. P | 0.998955 |

| Location | 11,104,838 – 11,104,929 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.68 |
| Shannon entropy | 0.73700 |
| G+C content | 0.59477 |
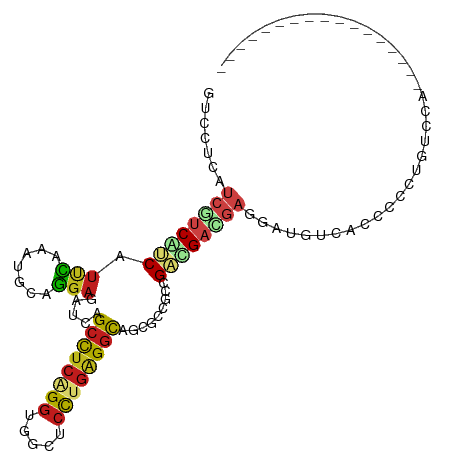
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.998955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
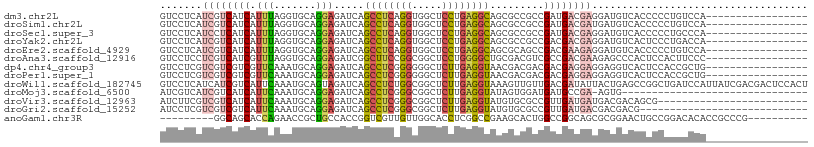
>dm3.chr2L 11104838 91 + 23011544 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCCGCCGAUGACGAGGAUGUCACCCCCUGUCCA----------------- .......((((((((.....(((((..(....).((((((((.....)))))))).)))))...))))))))(((((........))))).----------------- ( -39.40, z-score = -2.72, R) >droSim1.chr2L 10904171 91 + 22036055 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCCGCCGAUGACGAUGAUGUCACCCCCUGUCCA----------------- (.(.(((((((((((.....(((((..(....).((((((((.....)))))))).)))))...))))))))))).).)............----------------- ( -42.30, z-score = -3.96, R) >droSec1.super_3 6506208 91 + 7220098 GUCCUCAUCCUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCCGCCGAUGACGAGGAUGUCACCCCCUGCCCA----------------- .....(((((((.(((((..(((((..(....).((((((((.....)))))))).)))))...))))).)))))))..............----------------- ( -40.60, z-score = -2.99, R) >droYak2.chr2L 7500738 91 + 22324452 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCCGCCGACGACGAGGAUGUCACUCCCUGACCA----------------- ((((((.(((((........(((((..(....).((((((((.....)))))))).)))))...))))).))))))((((.....))))..----------------- ( -40.80, z-score = -2.95, R) >droEre2.scaffold_4929 12301478 91 - 26641161 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCAGCCGACGAAGAGGAUGUCACCCCCUGUCCA----------------- ((((((.(((((..(.....)((((..(....).((((((((.....)))))))).))))....))))).))))))...............----------------- ( -35.80, z-score = -1.96, R) >droAna3.scaffold_12916 12229841 91 + 16180835 GUCCUCCUCGUCAUCGUUUAGGUGCAGGAGAUCGGCUUCCGGCGGCUCCUGGGGCUGCGACGUCGCCGACGAAGAGCCCACUCCACUUCCC----------------- (..(((.(((((.......((.(.((((((.(((.(....).))))))))).).))(((....))).))))).)))..)............----------------- ( -32.00, z-score = 0.51, R) >dp4.chr4_group3 6455074 91 + 11692001 GUCCUCGUCGUCGUCGUUCAAAUGCAGGAGAUCAGCCUCGGGGGGCUCUUGAGGUAACGACGACGACGAGGAGGAGGUCACUCCACCGCUG----------------- .((((((((((((((((((.....(((((.....((((....)))))))))..).)))))))))))))))))((((....)))).......----------------- ( -48.40, z-score = -4.44, R) >droPer1.super_1 7946552 91 + 10282868 GUCCUCGUCGUCGUCGUUCAAAUGCAGGAGAUCAGCCUCGGGGGGCUCUUGAGGUAACGACGACGACGAGGAGGAGGUCACUCCACCGCUG----------------- .((((((((((((((((((.....(((((.....((((....)))))))))..).)))))))))))))))))((((....)))).......----------------- ( -48.40, z-score = -4.44, R) >droWil1.scaffold_182745 4807 108 + 6872 GUCCUCAUCAUCGUCAUUCAAAUGCAGUAGAUCAGCCUCUGGCGGCUCUUGAGGUAAAGUUGUUGACGAUAUUACUGAGCCGGCUGAUCCAUUAUCGACGACUCCACU ..........(((((.....((((.....((((((((......(((((....(((((.((((....)))).))))))))))))))))))))))...)))))....... ( -30.50, z-score = -1.19, R) >droMoj3.scaffold_6500 2929559 76 + 32352404 AUCGUCAUCGUCAUCAUUCAAAUGCAGGAGAUCAGCCUCGGGCGGCUCUUGAGGUAUAGUGGAUGAUGCCGA-AGUG------------------------------- .......(((.(((((((((....((((((....(((...)))..))))))........))))))))).)))-....------------------------------- ( -25.00, z-score = -1.33, R) >droVir3.scaffold_12963 12282364 83 - 20206255 AUCUUCGUCGUCAUCAUUCAAAUGCAGGAGAUCAGCCUCGGGCGGCUCUUGAGGUAUGUGCGCCGUUGAUGAUGACGACAGCG------------------------- ......(((((((((((.(((...((((((....(((...)))..)))))).(((......))).))))))))))))))....------------------------- ( -33.30, z-score = -2.37, R) >droGri2.scaffold_15252 6194808 80 - 17193109 AUCCUCGUCGUCGUCAUUCAAAUGCAGGAGAUCAGCCUCGGGCGGCUCUUGAGGUAUGUGCGCCGUUGAUGACGACGACG---------------------------- .....((((((((((((.(((...((((((....(((...)))..)))))).(((......))).)))))))))))))))---------------------------- ( -35.20, z-score = -2.85, R) >anoGam1.chr3R 6412858 89 + 53272125 ---------GGCAGCACCAGAACCGCUGCCACCGGUCGUUGUUGGCACCUCGGCCGAAGCACUGGCCGGCAGCGCGGAACUGCCGGACACACCGCCCG---------- ---------(((((..((.....(((((((.(((((.(((.(((((......)))))))))))))..))))))).))..)))))((.....)).....---------- ( -39.40, z-score = -1.07, R) >consensus GUCCUCAUCGUCAUCAUUCAAAUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGCGCCGCCGACGACGAGGAUGUCACCCCCUGUCCA_________________ .......((((((((.(((.......))).....((((((((.....)))))))).........)))))))).................................... (-16.39 = -16.71 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:34 2011