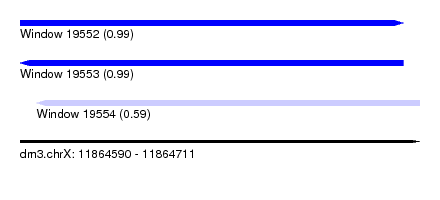
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,864,590 – 11,864,711 |
| Length | 121 |
| Max. P | 0.993549 |

| Location | 11,864,590 – 11,864,706 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.34194 |
| G+C content | 0.44612 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
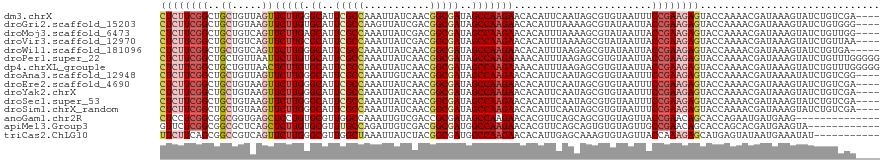
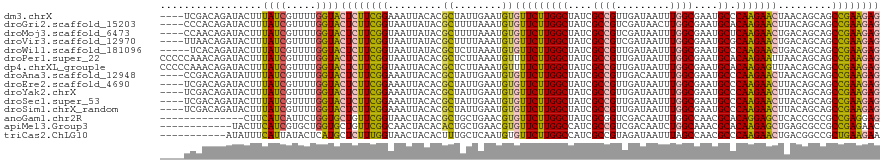
>dm3.chrX 11864590 116 + 22422827 CUCUUCGGCUGCUGUUAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.40, z-score = -3.71, R) >droGri2.scaffold_15203 11439756 116 + 11997470 CUCUUCGGCUGCUGUAAGUUCUUGUGCAUUCGCCAAGUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUGGG---- ((((((((((...((..(((((((.((..(((((...........)))))..)))))))))..)).....)))(((....))))))))))..(((....((((...))))..))).---- ( -34.90, z-score = -3.11, R) >droMoj3.scaffold_6473 12700697 116 + 16943266 CUCUUCGGCUGCUGUCAGUUCUUGAGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUUGG---- ((((((((((..(((..(((((((.((..(((((...........)))))..))))))))))))......)))(((....))))))))))..((((...((((...))))..))))---- ( -35.10, z-score = -3.79, R) >droVir3.scaffold_12970 8694234 116 + 11907090 CUCUUCGGCUGCUGUCAGUUCUUGCGCAUUCGCCAAAUUAUCGACGGCGAUAGCCAAGAACACAUUUAAAAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUUAA---- ((((((((((..(((..(((((((.((..(((((...........)))))..))))))))))))......)))(((....))))))))))......(((((((...)))).)))..---- ( -32.40, z-score = -3.13, R) >droWil1.scaffold_181096 10282967 115 + 12416693 CUCUUCGGCUGCUGUCAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUUAAGAGCGUAUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUGA----- ((((((((((.((....((((((((.(..(((((...........)))))..))))))))).......)))))(((....)))))))))).........((((...)))).....----- ( -32.30, z-score = -2.61, R) >droPer1.super_22 241742 120 + 1688296 CUCUUCGGCUGCUGUUAAUUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAAGAGCGUGUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUUUGGGGG (((((((((((((.((((((((((.((..(((((...........)))))..)))))))).....)))).)))).)......))))))))..(((((..((((...))))..)))))... ( -37.10, z-score = -3.44, R) >dp4.chrXL_group1e 2531154 120 - 12523060 CUCUUCGGCUGCUGUUAACUCUUGUGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAAAACAUUUAAGAGCGUGUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUUUGGGGG (((((((((((((.((((.(((((.((..(((((...........)))))..)))))))......)))).)))).)......))))))))..(((((..((((...))))..)))))... ( -35.10, z-score = -2.71, R) >droAna3.scaffold_12948 340548 116 + 692136 CUCUUCGGCUGCUGUUAGUUCUUGGGCAUUCGCCAAAUUGUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAAUAUCUGUCGG---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.60, z-score = -3.78, R) >droEre2.scaffold_4690 15435428 116 - 18748788 CUCUUCGGCUGCUGUAAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.40, z-score = -3.79, R) >droYak2.chrX 6933743 116 - 21770863 CUCUUCGGCUGCUGUAAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.40, z-score = -3.79, R) >droSec1.super_53 81180 116 + 190754 CUCUUCGGCUGCUGUAAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.40, z-score = -3.79, R) >droSim1.chrX_random 3263570 116 + 5698898 CUCUUCGGCUGCUGUAAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUUCCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA---- ((((((((.........((((((((.(..(((((...........)))))..)))))))))((((.(....).)))).....))))))))........(((((.......))))).---- ( -36.40, z-score = -3.79, R) >anoGam1.chr2R 23190333 106 + 62725911 CUCCUCGGCGGCGGUGAGCUCCUGUGCGUUGGCCAAAUUGUCGACCGCGAUAGCCAAGAACACGUUCAGCAGCGUGUAGUUACCGAACAGCACCAGAAUGAUGAAG-------------- ...((.(((..(((((((((..((((.((((((......))))))))))..))).....((((((......))))))..))))))....)).).))..........-------------- ( -31.00, z-score = 0.12, R) >apiMel3.Group3 11628386 108 + 12341916 GUUCUCGGCGGCGCUCAGCUCUUGUGCGUUUGCCAGAUUGUCGACGGCGAUGGCCAAGAACACGUUCAGCAGUGUGUAGUUGCCGAACAGCACCAGCACGAUGAAGUA------------ .((((((((((.(((..(.(((((.((..(((((...........)))))..))))))).)..((((.((((.......)))).))))))).)).)).))).)))...------------ ( -34.40, z-score = 0.73, R) >triCas2.ChLG10 899130 109 + 8806720 UUCUUCAGCGGCCGUCAGUUCUUGGGCGUUGGCUAAAUUAUCUACGGCGAUGGCCAAGAACACAUUGAGCAAAGUGUAGUUACCAAAGAGCAUGAGUAUAAUGAAAUAU----------- ...((((...((((((.((((((((.(((((.(............).))))).))))))))(((((......)))))............).))).))....))))....----------- ( -27.60, z-score = -0.72, R) >consensus CUCUUCGGCUGCUGUAAGUUCUUGGGCAUUCGCCAAAUUAUCAACGGCGAUAGCCAAGAACACAUUCAAUAGCGUGUAAUUACCGAAGAGUACCAAAACGAUAAAGUAUCUGUCGA____ ((((((((..((.....))(((((.((..(((((...........)))))..))))))).......................)))))))).............................. (-22.38 = -22.92 + 0.54)
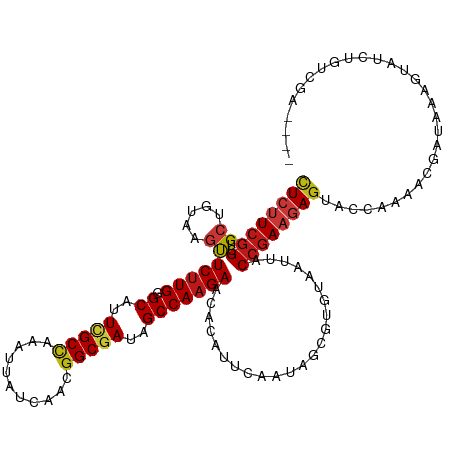
| Location | 11,864,590 – 11,864,706 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.34194 |
| G+C content | 0.44612 |
| Mean single sequence MFE | -34.69 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11864590 116 - 22422827 ----UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUAACAGCAGCCGAAGAG ----....((((.((......))))))...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -34.60, z-score = -2.87, R) >droGri2.scaffold_15203 11439756 116 - 11997470 ----CCCACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAACUUGGCGAAUGCACAAGAACUUACAGCAGCCGAAGAG ----.(((..((((...))))....)))..(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........))))))))) ( -36.30, z-score = -3.83, R) >droMoj3.scaffold_6473 12700697 116 - 16943266 ----CCAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCUCAAGAACUGACAGCAGCCGAAGAG ----((((..((((...))))...))))..(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........))))))))) ( -37.60, z-score = -3.91, R) >droVir3.scaffold_12970 8694234 116 - 11907090 ----UUAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCGCAAGAACUGACAGCAGCCGAAGAG ----....((((.((......))))))...(((((((((........((........))(((((((((..((((((.........))))))..)).)))))))........))))))))) ( -35.10, z-score = -2.80, R) >droWil1.scaffold_181096 10282967 115 - 12416693 -----UCACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUCUUAAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUGACAGCAGCCGAAGAG -----...((((.((......))))))...(((((((((........((........))(((((((((..((((((.........))))))..).))))))))........))))))))) ( -35.10, z-score = -3.17, R) >droPer1.super_22 241742 120 - 1688296 CCCCCAAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAAUUAACAGCAGCCGAAGAG ...(((((..((((...))))..)))))..(((((((((........(((.((((.....((((((((..((((((.........))))))..)).)))))))))).))).))))))))) ( -36.10, z-score = -4.06, R) >dp4.chrXL_group1e 2531154 120 + 12523060 CCCCCAAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAGUUAACAGCAGCCGAAGAG ...(((((..((((...))))..)))))..(((((((((........(((.((((.....((((((((..((((((.........))))))..)).)))))))))).))).))))))))) ( -35.80, z-score = -3.60, R) >droAna3.scaffold_12948 340548 116 - 692136 ----CCGACAGAUAUUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGACAAUUUGGCGAAUGCCCAAGAACUAACAGCAGCCGAAGAG ----((((..((((...))))...))))..((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -36.80, z-score = -3.63, R) >droEre2.scaffold_4690 15435428 116 + 18748788 ----UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCGAAGAG ----....((((.((......))))))...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -34.60, z-score = -2.87, R) >droYak2.chrX 6933743 116 + 21770863 ----UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCGAAGAG ----....((((.((......))))))...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -34.60, z-score = -2.87, R) >droSec1.super_53 81180 116 - 190754 ----UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCGAAGAG ----....((((.((......))))))...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -34.60, z-score = -2.87, R) >droSim1.chrX_random 3263570 116 - 5698898 ----UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCGAAGAG ----....((((.((......))))))...((((((((.....(((..(....)..)))(((((((((..((((((.........))))))..).)))))))).........)))))))) ( -34.60, z-score = -2.87, R) >anoGam1.chr2R 23190333 106 - 62725911 --------------CUUCAUCAUUCUGGUGCUGUUCGGUAACUACACGCUGCUGAACGUGUUCUUGGCUAUCGCGGUCGACAAUUUGGCCAACGCACAGGAGCUCACCGCCGCCGAGGAG --------------........(((((((((.((((((((.(.....).))))))))(.(((((((((....))((((((....))))))......))))))).)...).)))).)))). ( -33.70, z-score = -0.49, R) >apiMel3.Group3 11628386 108 - 12341916 ------------UACUUCAUCGUGCUGGUGCUGUUCGGCAACUACACACUGCUGAACGUGUUCUUGGCCAUCGCCGUCGACAAUCUGGCAAACGCACAAGAGCUGAGCGCCGCCGAGAAC ------------...(((.(((.((.((((((((((((((.........))))))))..(((((((((....((((.........))))....)).)))))))..)))))))))))))). ( -41.40, z-score = -2.54, R) >triCas2.ChLG10 899130 109 - 8806720 -----------AUAUUUCAUUAUACUCAUGCUCUUUGGUAACUACACUUUGCUCAAUGUGUUCUUGGCCAUCGCCGUAGAUAAUUUAGCCAACGCCCAAGAACUGACGGCCGCUGAAGAA -----------..............(((.((....((((....((((..........)))).....))))..(((((..........((....))..........))))).))))).... ( -19.45, z-score = 0.56, R) >consensus ____UCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUGACAGCAGCCGAAGAG .................((((.....))))((((((((.........((........))(((((((((....((((.........))))....)).))))))).........)))))))) (-24.78 = -24.48 + -0.30)
| Location | 11,864,595 – 11,864,711 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Shannon entropy | 0.39949 |
| G+C content | 0.44527 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
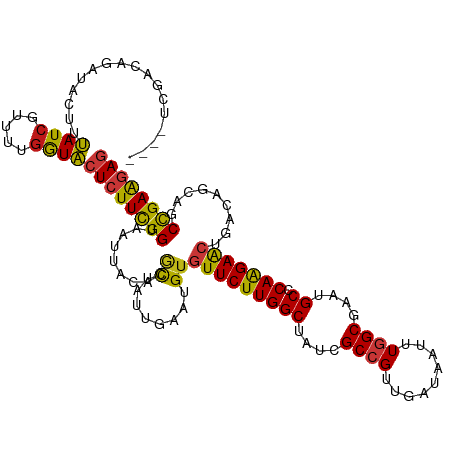
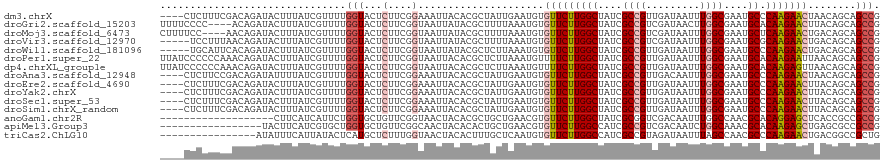
>dm3.chrX 11864595 116 - 22422827 ----CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUAACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -31.70, z-score = -2.50, R) >droGri2.scaffold_15203 11439761 116 - 11997470 UUUUCCCC----ACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAACUUGGCGAAUGCACAAGAACUUACAGCAGCCG ......((----(..((((...))))....))).......((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))) ( -26.00, z-score = -1.23, R) >droMoj3.scaffold_6473 12700702 116 - 16943266 CUUUUCC----AACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCUCAAGAACUGACAGCAGCCG .....((----((..((((...))))...)))).......((((........((........))(((((((((..((((((.........))))))..)).)))))))........)))) ( -27.40, z-score = -1.43, R) >droVir3.scaffold_12970 8694239 115 - 11907090 -----UCCUUUAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUUUUAAAUGUGUUCUUGGCUAUCGCCGUCGAUAAUUUGGCGAAUGCGCAAGAACUGACAGCAGCCG -----..........((((...)))).....(((...(((((((......((((........))))(((((((..((((((.........))))))..)).)))))))))).))..))). ( -26.80, z-score = -0.76, R) >droWil1.scaffold_181096 10282972 115 - 12416693 -----UGCAUUCACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUAUACGCUCUUAAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUGACAGCAGCCG -----(((......(((((((..(....)..)))).)))(((((......((((........))))(((((((..((((((.........))))))..).)))))))))))..))).... ( -27.60, z-score = -0.98, R) >droPer1.super_22 241747 120 - 1688296 UUAUCCCCCCAAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAAUUAACAGCAGCCG ........(((((..((((...))))..)))))((((....)))).......(((.((((.....((((((((..((((((.........))))))..)).)))))))))).)))..... ( -26.20, z-score = -1.51, R) >dp4.chrXL_group1e 2531159 120 + 12523060 UUAUCCCCCCAAACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUCUUAAAUGUUUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCACAAGAGUUAACAGCAGCCG ........(((((..((((...))))..)))))((((....)))).......(((.((((.....((((((((..((((((.........))))))..)).)))))))))).)))..... ( -25.90, z-score = -1.09, R) >droAna3.scaffold_12948 340553 116 - 692136 ----CUCUUCCGACAGAUAUUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGACAAUUUGGCGAAUGCCCAAGAACUAACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -32.20, z-score = -2.56, R) >droEre2.scaffold_4690 15435433 116 + 18748788 ----CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -31.70, z-score = -2.52, R) >droYak2.chrX 6933748 116 + 21770863 ----CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -31.70, z-score = -2.52, R) >droSec1.super_53 81185 116 - 190754 ----CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -31.70, z-score = -2.52, R) >droSim1.chrX_random 3263575 116 - 5698898 ----CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGAAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUUACAGCAGCCG ----...((((((.(((((((..(....)..)))).))))))))).......(((.(((.....(((((((((..((((((.........))))))..).))))))))...))).))).. ( -31.70, z-score = -2.52, R) >anoGam1.chr2R 23190338 101 - 62725911 -------------------CUUCAUCAUUCUGGUGCUGUUCGGUAACUACACGCUGCUGAACGUGUUCUUGGCUAUCGCGGUCGACAAUUUGGCCAACGCACAGGAGCUCACCGCCGCCG -------------------............(((((.((((((((.(.....).))))))))(.(((((((((....))((((((....))))))......))))))).)...).)))). ( -31.60, z-score = -0.92, R) >apiMel3.Group3 11628391 103 - 12341916 -----------------UACUUCAUCGUGCUGGUGCUGUUCGGCAACUACACACUGCUGAACGUGUUCUUGGCCAUCGCCGUCGACAAUCUGGCAAACGCACAAGAGCUGAGCGCCGCCG -----------------........((.((.((((((((((((((.........))))))))..(((((((((....((((.........))))....)).)))))))..)))))))))) ( -37.40, z-score = -1.78, R) >triCas2.ChLG10 899135 104 - 8806720 ----------------AUAUUUCAUUAUACUCAUGCUCUUUGGUAACUACACUUUGCUCAAUGUGUUCUUGGCCAUCGCCGUAGAUAAUUUAGCCAACGCCCAAGAACUGACGGCCGCUG ----------------..................((....((((....((((..........)))).....))))..(((((..........((....))..........))))).)).. ( -17.45, z-score = 0.60, R) >consensus ____CUCUUUCGACAGAUACUUUAUCGUUUUGGUACUCUUCGGUAAUUACACGCUAUUGAAUGUGUUCUUGGCUAUCGCCGUUGAUAAUUUGGCGAAUGCCCAAGAACUGACAGCAGCCG ...............................(((...(....).....................(((((((((....((((.........))))....)).)))))))........))). (-16.18 = -16.49 + 0.31)
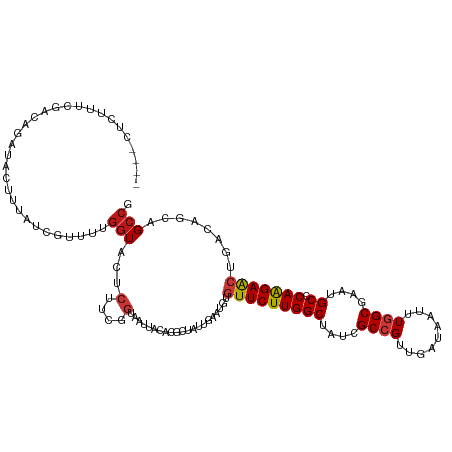
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:49 2011