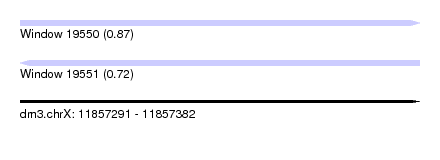
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,857,291 – 11,857,382 |
| Length | 91 |
| Max. P | 0.867164 |

| Location | 11,857,291 – 11,857,382 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Shannon entropy | 0.37212 |
| G+C content | 0.30272 |
| Mean single sequence MFE | -18.11 |
| Consensus MFE | -8.77 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867164 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11857291 91 + 22422827 AUCAUUAAAUAUUCC------CAGACUAAUGCGGAAUAUUUAAGGACUUAUC---GUUUGUGAA-AUGAUAUAUCUCUAUUUUCCGGUAAAUGUUCACUAU (((.(((((((((((------((......)).)))))))))))(((..((((---(((.....)-))))))..))).........)))............. ( -18.70, z-score = -2.02, R) >droSim1.chrX_random 3255299 90 + 5698898 AUCGUUAAAUAUUCC------AAAACUAAUGCGGAAUAUUUAAGGACU-AUC---GUGUGUGAA-AUGAUAUAUCUCUAUUUUCCGGUAAAUGUUCACUAU .((.(((((((((((------...........))))))))))).))..-...---....(((((-...((.((((..........)))).)).)))))... ( -17.00, z-score = -1.62, R) >droSec1.super_53 73890 93 + 190754 AUCGUUAAAUAUUCC------AAAACUAAUGCGGAAUAUUUAAGGACU-AUCAUCGUGUGUGAA-AUGAUAUAUCUCUAUUUUCCGGUAAAUGUUCACUAU .((.(((((((((((------...........))))))))))).))..-..........(((((-...((.((((..........)))).)).)))))... ( -17.00, z-score = -1.42, R) >droEre2.scaffold_4690 15428194 90 - 18748788 AUCGUUAAAUAUUCC------AAGACUACUGGGGAAUAUUUAAGGACU-AUC---GUGUGUGAA-AUGAUACACCUCUAUUUUCCGGUAAAUGUUCACUAU .((.(((((((((((------.((....))..))))))))))).))..-...---((((((...-...))))))........................... ( -19.50, z-score = -1.22, R) >droAna3.scaffold_12948 334388 90 + 692136 AUCAUUAAAUAUUCCC-----ACUACAAUCGGGAAAUAUUUAAGGAUU-AUC---GCAUGAAUU--GAAUACAUCUCUACUUUCCGGUAAAUGUUCACUAU .((.((((((((((((-----.........))).))))))))).))((-(((---(...(((..--((.......))....)))))))))........... ( -13.30, z-score = -0.71, R) >droWil1.scaffold_181096 10274481 96 + 12416693 AUCGUUAAAUAUUCCCAAAGCAAAAUUGCUA-GGAAUAUUUAAGGACU-AUC---GUGAUCAUUUAAAAUAAAUCUCUACUUUCCGGUAAAUGUUCACUAU .((.(((((((((((...((((....)))).-))))))))))).))..-...---((((.((((((....(((.......)))....)))))).))))... ( -23.80, z-score = -4.89, R) >droVir3.scaffold_12970 8686853 83 + 11907090 AUCGUUAAAUAUUC----------AUCACU---GAAUAUUUAAGGACU-AUC---GUGGGUAUU-AUAAAAAAUCUCUACUUUCCGGUAAAUGUUCACUCU .((.((((((((((----------(....)---)))))))))).))..-...---(((..((((-............((((....))))))))..)))... ( -17.20, z-score = -3.35, R) >droMoj3.scaffold_6473 12693730 83 + 16943266 AUCGUUAAAUAUUC----------AUGACU---GAAUAUUUAAGGACU-AUC---GUGAGUAUU-AUCAAAAAUCUCUACUUUCCGGUAAAUGUUCACUCU .((.((((((((((----------(....)---)))))))))).))..-...---(((((((((-(((.................))).)))))))))... ( -16.63, z-score = -2.55, R) >droGri2.scaffold_15203 11432105 83 + 11997470 AUCGUUAAAUAUUC----------AAAACU---GAAUAUUUAAGGACU-AUC---GUGAAUAUU-AUGAUAAAUCUCUACUUUCCGGUAAAUGUUCACUCU .((.((((((((((----------(....)---)))))))))).))..-...---(((((((((-..((......))((((....)))))))))))))... ( -19.90, z-score = -4.55, R) >consensus AUCGUUAAAUAUUCC______AA_ACUACUG_GGAAUAUUUAAGGACU_AUC___GUGUGUAUU_AUGAUAAAUCUCUACUUUCCGGUAAAUGUUCACUAU .((.(((((((((((.................))))))))))).))....................................................... ( -8.77 = -8.96 + 0.19)



| Location | 11,857,291 – 11,857,382 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Shannon entropy | 0.37212 |
| G+C content | 0.30272 |
| Mean single sequence MFE | -17.59 |
| Consensus MFE | -9.11 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720174 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11857291 91 - 22422827 AUAGUGAACAUUUACCGGAAAAUAGAGAUAUAUCAU-UUCACAAAC---GAUAAGUCCUUAAAUAUUCCGCAUUAGUCUG------GGAAUAUUUAAUGAU ...(((((.((((......)))).((......))..-)))))....---.....(((.(((((((((((.((......))------))))))))))).))) ( -16.70, z-score = -1.51, R) >droSim1.chrX_random 3255299 90 - 5698898 AUAGUGAACAUUUACCGGAAAAUAGAGAUAUAUCAU-UUCACACAC---GAU-AGUCCUUAAAUAUUCCGCAUUAGUUUU------GGAAUAUUUAACGAU ...(((...((((......)))).(((((.....))-))).)))..---...-.(((.((((((((((((.........)------))))))))))).))) ( -15.50, z-score = -1.80, R) >droSec1.super_53 73890 93 - 190754 AUAGUGAACAUUUACCGGAAAAUAGAGAUAUAUCAU-UUCACACACGAUGAU-AGUCCUUAAAUAUUCCGCAUUAGUUUU------GGAAUAUUUAACGAU ...((((....))))...............((((((-(........))))))-)(((.((((((((((((.........)------))))))))))).))) ( -16.40, z-score = -1.60, R) >droEre2.scaffold_4690 15428194 90 + 18748788 AUAGUGAACAUUUACCGGAAAAUAGAGGUGUAUCAU-UUCACACAC---GAU-AGUCCUUAAAUAUUCCCCAGUAGUCUU------GGAAUAUUUAACGAU ...(((((....((((..........))))......-)))))....---...-.(((.(((((((((((..((....)).------))))))))))).))) ( -19.10, z-score = -1.83, R) >droAna3.scaffold_12948 334388 90 - 692136 AUAGUGAACAUUUACCGGAAAGUAGAGAUGUAUUC--AAUUCAUGC---GAU-AAUCCUUAAAUAUUUCCCGAUUGUAGU-----GGGAAUAUUUAAUGAU ...(((((..(((((......)))))((.....))--..)))))..---...-.(((.(((((((((((((.......).-----)))))))))))).))) ( -16.90, z-score = -0.80, R) >droWil1.scaffold_181096 10274481 96 - 12416693 AUAGUGAACAUUUACCGGAAAGUAGAGAUUUAUUUUAAAUGAUCAC---GAU-AGUCCUUAAAUAUUCC-UAGCAAUUUUGCUUUGGGAAUAUUUAACGAU ...((((.((((((.....((((((....)))))))))))).))))---...-.(((.(((((((((((-(((((....))))..)))))))))))).))) ( -25.50, z-score = -4.24, R) >droVir3.scaffold_12970 8686853 83 - 11907090 AGAGUGAACAUUUACCGGAAAGUAGAGAUUUUUUAU-AAUACCCAC---GAU-AGUCCUUAAAUAUUC---AGUGAU----------GAAUAUUUAACGAU ...(((.....(((..(((((((....))))))).)-))....)))---...-.(((.((((((((((---(....)----------)))))))))).))) ( -15.30, z-score = -2.43, R) >droMoj3.scaffold_6473 12693730 83 - 16943266 AGAGUGAACAUUUACCGGAAAGUAGAGAUUUUUGAU-AAUACUCAC---GAU-AGUCCUUAAAUAUUC---AGUCAU----------GAAUAUUUAACGAU ...((((....(((.((((((.......)))))).)-))...))))---...-.(((.((((((((((---(....)----------)))))))))).))) ( -17.40, z-score = -2.35, R) >droGri2.scaffold_15203 11432105 83 - 11997470 AGAGUGAACAUUUACCGGAAAGUAGAGAUUUAUCAU-AAUAUUCAC---GAU-AGUCCUUAAAUAUUC---AGUUUU----------GAAUAUUUAACGAU ...(((((..(((((......)))))((....))..-....)))))---...-.(((.((((((((((---......----------)))))))))).))) ( -15.50, z-score = -2.10, R) >consensus AUAGUGAACAUUUACCGGAAAGUAGAGAUUUAUCAU_AAUACACAC___GAU_AGUCCUUAAAUAUUCC_CAGUAGU_UU______GGAAUAUUUAACGAU ...((((....)))).......................................(((.(((((((((((.................))))))))))).))) ( -9.11 = -9.00 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:46 2011