
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,830,272 – 11,830,365 |
| Length | 93 |
| Max. P | 0.701332 |

| Location | 11,830,272 – 11,830,365 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Shannon entropy | 0.55745 |
| G+C content | 0.53026 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
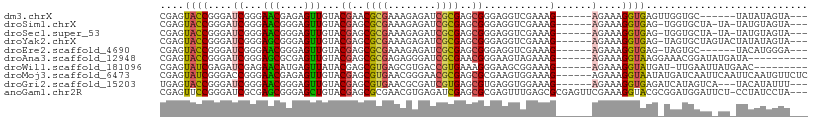
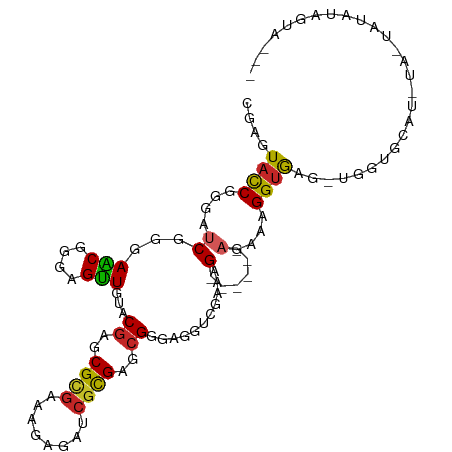
>dm3.chrX 11830272 93 - 22422827 CGAGUACCGGGAUCGGGAACGAGAGUUGUACGAACGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG------AGAAAGGUGAGUUGGUGC------UAUAUAGUA--- ..((((((((..((((.(((....))).).((..(((((......)))))..)).....((....)------)......))).)))))))------)........--- ( -25.40, z-score = -2.10, R) >droSim1.chrX 9108569 96 - 17042790 CGAGUACCGGGAUCGGGAACGGGAGUUGUACGAGCGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG------AGAAAGGUGAG-UGGUGCUA-UA-UAUGUAGUA--- ..(((((((...((((.(((....))).).((..(((((......)))))..)).....((....)------)......))).-))))))).-..-.........--- ( -25.10, z-score = -1.75, R) >droSec1.super_53 47455 96 - 190754 CGAGUACCGGGAUCGGGAACGGGAGUUGUACGAGCGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG------AGAAAGGUGAG-UGGUGCUA-UA-UAUGUAGUA--- ..(((((((...((((.(((....))).).((..(((((......)))))..)).....((....)------)......))).-))))))).-..-.........--- ( -25.10, z-score = -1.75, R) >droYak2.chrX 6900950 98 + 21770863 CGAGUACCGGGAUCGGGAGCGGGAGUUGUACGAGCGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG------AGAAAGGUGAG-UAGUGCUAGUACUAUAUAGUA--- ....((((....((.(.(((....))).).((..(((((......)))))..)).))..((....)------)....)))).(-(((((....))))))......--- ( -24.50, z-score = -0.87, R) >droEre2.scaffold_4690 15401639 92 + 18748788 CGAGUACCGGGAUCGGGAACGGGAGUUGUACGAGCGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG------AGAAAGGUGAG-UAGUGC------UACAUGGGA--- ......(((...(((....)))..((.(((((..(((((......)))))..)......((....)------)..........-..))))------.)).)))..--- ( -22.60, z-score = -0.85, R) >droAna3.scaffold_12948 307630 92 - 692136 CGAGUACCGGGAUCGGGAGCGCGAGUUGUACGAGCGCGAGAGGGAUCGCGAACGGGAAGUAGAAAG------AGAAAGGUAAGGAAACGGAUAUGAUA---------- ....((((....((.(.(((....))).).((..(((((......)))))..))...........)------)....)))).(....)..........---------- ( -21.50, z-score = -1.73, R) >droWil1.scaffold_181096 10234983 92 - 12416693 CGAGUAUCGAGAUCGAGAACAUGAGUUAUACGAGCGUGAGCGUGACCGUGAAAGGGAAGCGGAAAG------AGAAAGGUAUGAU-UUGAAUUAUGAAC--------- ...((.(((....)))..))..(((((((((..((....))....((((.........))))....------......)))))))-))...........--------- ( -19.70, z-score = -2.61, R) >droMoj3.scaffold_6473 12658157 102 - 16943266 CGAGUAUCGGGACCGGGAACGAGAGUUGUACGAGCGUGAACGGGAACGCGAGCGCGAAGUGGAAAG------AGAAAGGUAAUAUGAUCAAUUCAAUUCAAUGUUCUC ((.....)).....(((((((.(((((((((..((((...((....))...))))...))).....------.....(((......)))....))))))..))))))) ( -22.30, z-score = 0.41, R) >droGri2.scaffold_15203 11398880 96 - 11997470 UGAGUACCGGGAUCGGGAACGGGAGUUGUACGAGCGUGAACGCGAUCGUGAGCGUGAGGUGGAAAG------AGAAAGGUGAGAUCAUAGUCA---UACAUAUUU--- (((.((((..((((.(.(((....))).)....(((....))))))).(.(.(....).).)....------.....))))...)))......---.........--- ( -20.70, z-score = 0.04, R) >anoGam1.chr2R 23164105 104 - 62725911 CGAGUUCCGGGAUCGCGAGCGGGAGCUGUACGAGCGCGAACGUGAGAUCGAGCGCGAGUUUGAGCGCGAGUUCGAAAGGUACGCGGAUGGAUUCU-CCUAUCCUA--- .(((.((((...((((.(((....)))((((..((((...(....).((((((....)))))))))).....(....))))))))).))))..))-)........--- ( -39.60, z-score = -0.27, R) >consensus CGAGUACCGGGAUCGGGAACGGGAGUUGUACGAGCGCGAAAGAGAUCGCGAGCGGGAGGUCGAAAG______AGAAAGGUGAG_UGGUGCAU_UA_UAUAUAGUA___ ....((((.........(((....)))...((..((((........))))..)).......................))))........................... (-12.64 = -12.00 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:42 2011