| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,814,219 – 11,814,390 |
| Length | 171 |
| Max. P | 0.995714 |

| Location | 11,814,219 – 11,814,390 |
|---|---|
| Length | 171 |
| Sequences | 5 |
| Columns | 179 |
| Reading direction | reverse |
| Mean pairwise identity | 68.98 |
| Shannon entropy | 0.54413 |
| G+C content | 0.34581 |
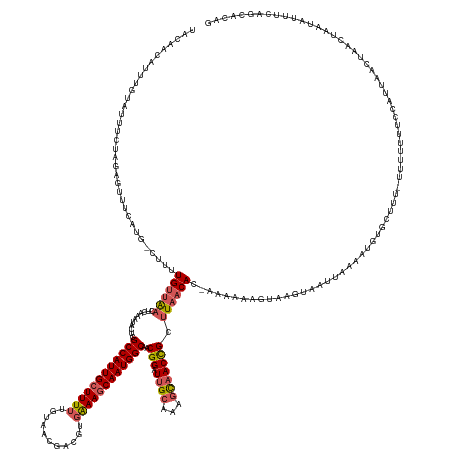
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
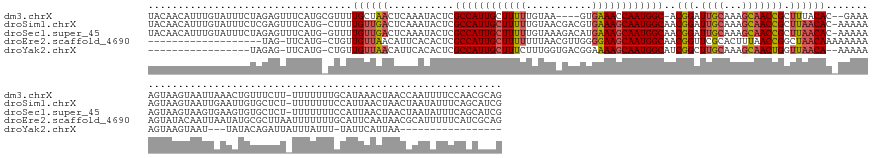
>dm3.chrX 11814219 171 - 22422827 UACAACAUUUGUAUUUCUAGAGUUUCAUGCGUUUUGCUAACUCAAAUACUCGCCAUUGCUUUUUGUAA----GUGAAACCAAUGGC-ACGGAUUGCAAAGCAACCGCUUUACAC--GAAAAGUAAGUAAUUAAACUGUUUCUU-UUUUUUUGCAUAAACUAACCAAUUUUCCAACGCAG ..........((((((...(((((....((.....)).)))))))))))..(((((((.((((.....----..)))).)))))))-..(((.((.(((((....))))).))(--((((((.(((.(((......))).)))-.))))))).................)))....... ( -32.20, z-score = -0.62, R) >droSim1.chrX 7801467 176 + 17042790 UACAACAUUUGUAUUUCUCGAGUUUCAUG-CUUUUGUUGACUCAAAUACUCGCCAUUGCUUUUUGUAACGACGUGAAAGCAAUGGCAACGGAUUGCAAAGCAACCGCUUAACAC-AAAAAAGUAAGUAAUUGAAUUGUGCUCU-UUUUUUUCCAUUAACUAACUAAUAUUUCAGCAUCG ..........((((((...(((((.((..-....))..)))))))))))..(((((((((((((((....))).))))))))))))....(((.((.((((....)))).....-.....(((.((((((.(((..(......-..)..))).))).))).))).........))))). ( -37.30, z-score = -1.49, R) >droSec1.super_45 44114 176 - 245362 UACAACAUUUGUAUUUCUAGAGUUUCAUG-GUUUUGUUGACUCAAAUACUCGCCAUUGCUUUUUGUAAAGACAUGAAAGCAAUGGCAACGGAUUGCAAAGCAACCGCUUAACAC-AAAAAAGUAAGUAAGUGAAGUGUGCUCU-UUUUUUUCCAUUAACUAACUAAUAUUUCAGCAUCG .......((((((.(((..((((.....(-(((.....)))).....))))(((((((((((((((....))).))))))))))))...))).))))))((....(((((..((-......))...)))))(((((((.....-.....................))))))).)).... ( -41.17, z-score = -1.95, R) >droEre2.scaffold_4690 14847035 158 - 18748788 -------------------UAG-UUCAUG-CUGUUGUUAACAUUCACACUCGCCAUUGCUUUUUUUAACGUUGGGGAAGCAAUGGCAACGGUUCGCACUUUAACCGGCUAACAAAAAAAAAGUAUACAAUUAAUAUGCGCUUAAUUUUUUUGCAUUCAAUAACGCAUUUUUCAUCGCAG -------------------...-...(((-(.((((((.............((((((((((((((.......))))))))))))))..(((((........))))).....((((((((..(((((.......)))))......)))))))).....))))))))))............ ( -35.60, z-score = -1.92, R) >droYak2.chrX 1868954 137 - 21770863 -----------------UAGAG-UUCAUG-CUGUUGUUAACAUUCACACUCGCCAUUGCUUUCUUUGGUGACGGAAAAGCAAUGGCAUCGGCUUGCAAAGCAACUGGUUAACA--AAAAAAGUAAGUAAU---UAUACAGAUUAUUUAUUU-UAUUCAUUAA----------------- -----------------..(((-(.....-...((((((((..........(((((((((((.((((....)))))))))))))))..(((.((((...))))))))))))))--)..((((((((((((---(.....))))))))))))-))))).....----------------- ( -43.90, z-score = -5.58, R) >consensus UACAACAUUUGUAUUUCUAGAGUUUCAUG_CUUUUGUUAACUCAAAUACUCGCCAUUGCUUUUUGUAACGACGUGAAAGCAAUGGCAACGGAUUGCAAAGCAACCGCUUAACAC_AAAAAAGUAAGUAAUUAAAAUGUGCUUU_UUUUUUUCCAUUAACUAACUAAUAUUUCAGCACAG ..................................((((((...........((((((((((((...........))))))))))))..(((.((((...))))))).)))))).................................................................. (-21.14 = -21.82 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:39 2011