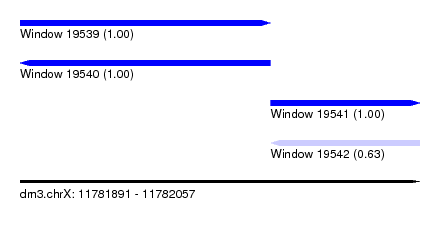
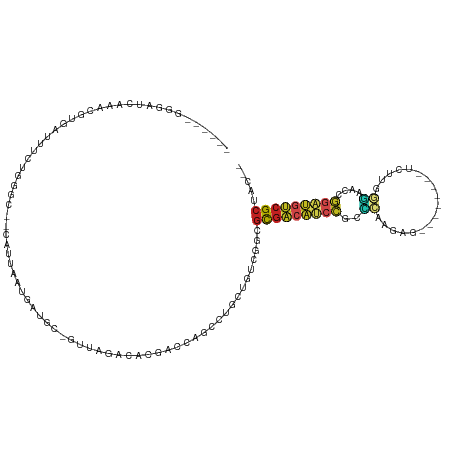
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,781,891 – 11,782,057 |
| Length | 166 |
| Max. P | 0.999598 |

| Location | 11,781,891 – 11,781,995 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 50.59 |
| Shannon entropy | 0.98166 |
| G+C content | 0.50312 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -13.62 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
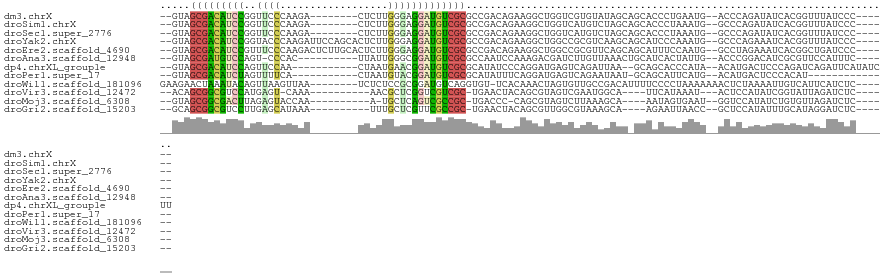
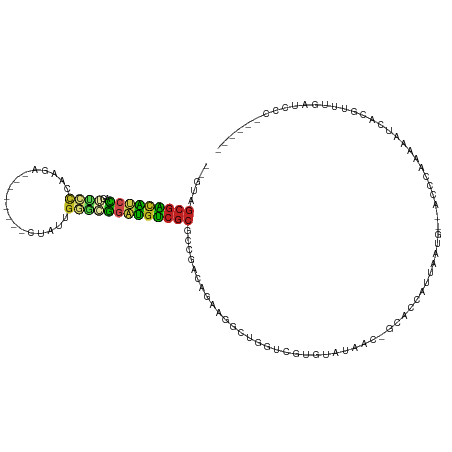
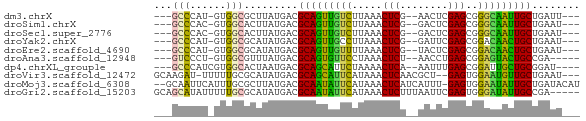
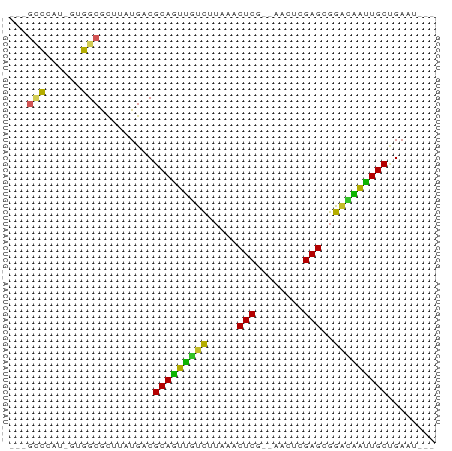
>dm3.chrX 11781891 104 + 22422827 --GUAGCGACAUCCGGUUCCCAAGA--------CUCUUGGGAGGAUGUCGCGCCGACAGAAGGCUGGUCGUGUAUAGCAGCACCCUGAAUG--ACCCAGAUAUCACGGUUUAUCCC------ --...(((((((((...(((((((.--------..))))))))))))))))((((...((...((((((((......(((....))).)))--).))))...)).)))).......------ ( -39.30, z-score = -2.68, R) >droSim1.chrX 9082224 104 + 17042790 --GUAGCGACAUCCGGUUCCCAAGA--------CUCUUGGGAGGAUGUCGCGCCGACAGAAGGCUGGUCAUGUCUAGCAGCACCCUAAAUG--GCCCAGAUAUCACGGUUUAUCCC------ --...(((((((((...(((((((.--------..))))))))))))))))((((...((...((((((((...(((.......))).)))--).))))...)).)))).......------ ( -38.20, z-score = -2.28, R) >droSec1.super_2776 378 104 + 2999 --GUAGCGACAUCCGGUUCCCAAGA--------CUCUUGGGAGGAUGUCGCGCCGACAGAAGGCUGGUCAUGUCUAGCAGCACCCUAAAUG--GCCCAGAUAUCACGGUUUAUCCC------ --...(((((((((...(((((((.--------..))))))))))))))))((((...((...((((((((...(((.......))).)))--).))))...)).)))).......------ ( -38.20, z-score = -2.28, R) >droYak2.chrX 6854945 112 - 21770863 --GUAGCGACAUCCGGUACCCAAGAUUCCAGCACUCUUGGGAGGAUGUCGCGCCGACAGAAGGCUGGCCGCGUCAAGCAGCAUCCCAAAUG--GCCCAGAAAUCACGGUUUAUCCC------ --...(((((((((....(((((((.........))))))).)))))))))((((...((...((((((((.....)).(.....)....)--).))))...)).)))).......------ ( -39.70, z-score = -2.12, R) >droEre2.scaffold_4690 15331120 112 - 18748788 --GUAGCGACAUCCGUUUCCCAAGACUCUUGCACUCUUGGGAGGAUGUCGCGCCGACAGAAGGCUGGCCGCGUUCAGCAGCAUUUCCAAUG--GCCUAGAAAUCACGGCUGAUCCC------ --...(((((((((...((((((((.........)))))))))))))))))((((...((...((((((((.....)).(......)...)--))).))...)).)))).......------ ( -43.20, z-score = -2.54, R) >droAna3.scaffold_12948 293408 101 - 692136 --GUAGCGAUGUCCAGU-CCCAC----------UUAUUGGGCGGAUGUCGCGCCAAUCCAAAGACGAUCUUGUUAAACUGCAUCACUAUUG--ACCCGGACAUCGCGUUCCAUUUC------ --...(((((((((.((-(....----------..((((((((.....))).))))).....))).....(((......))).........--....)))))))))..........------ ( -29.90, z-score = -2.10, R) >dp4.chrXL_group1e 8000602 105 - 12523060 --GUAGCGACAUCCAGUUCCAA-----------CUAAUGAACGGAUGUCGCGCAUAUCCCAGGAUGAGUCAGAUUAA--GCAGCACCCAUA--ACAUGACUCCCAGAUCAGAUUCAUAUCUU --...(((((((((.((((...-----------.....))))))))))))).........((((((((((.((((..--(.((((......--...)).)).)..)))).))))))).))). ( -29.70, z-score = -3.79, R) >droPer1.super_17 467546 92 - 1930428 --GUAGCGACAUCUAGUUUUCA-----------CUAAUGUACGGAUGUCGCGCAUAUUUCAGGAUGAGUCAGAAUAAU-GCAGCAUUCAUG--ACAUGACUCCCACAU-------------- --...(((((((((.((...((-----------....)).)))))))))))..........((..((((((.....((-(.......))).--...))))))))....-------------- ( -23.70, z-score = -1.49, R) >droWil1.scaffold_181096 10677489 107 - 12416693 GAAGAACUAAAUACAGUUAAGUUAA--------UCUCUCCGCGGAUGUCAGGUGU-UCACAAACUAGUGUUGCCGACAUUUUCCCCUAAAAAAACUCUAAAAUUGUCAUUCAUCUC------ ((.(((......((((((.((((..--------.......(..((((((.((((.-.(((......))).))))))))))..).........))))....))))))..))).))..------ ( -15.47, z-score = -0.36, R) >droVir3.scaffold_12472 135760 95 + 763072 --ACAGCGGCGUCCAUGAGU-CAAA----------AACGCUCGGUCGUCGC-UGAACUACAGCGUAGUCGAAUGGCA----UUCAUAAAU---ACUCCAUAUCGGUAUUAGAUCUC------ --.((((((((.((..((((-....----------...)))))).))))))-)).(((((...)))))(((((((..----.........---...)))).)))............------ ( -25.84, z-score = -1.78, R) >droMoj3.scaffold_6308 1937850 95 - 3356042 --GUAGCGGCGACUUAGAGUACCAA----------A-UGCUCAGUCGCCGC-UGACCC-CAGCGUAGUCUUAAAGCA----AAUAGUGAAU--GGUCCAUAUCUGUGUUAGAUCUC------ --.(((((((((((..(((((....----------.-))))))))))))))-))....-.......((((....(((----.(((..((..--..))..))).)))...))))...------ ( -28.50, z-score = -2.58, R) >droGri2.scaffold_15203 9922994 97 - 11997470 --GCAGCGGCGUCCUUGAGCAUAAA----------UUUGCUCGUUCGCCGC-UGAACUACAGCGUUGGCGUAAAGCA----AGAAUUAACC--GCUCCAUAUUUGCAUAGGAUCUC------ --.((((((((....((((((....----------..))))))..))))))-))..(((..(((.(((.....(((.----..........--))))))....))).)))......------ ( -30.30, z-score = -1.58, R) >consensus __GUAGCGACAUCCAGUUCCCAAGA________CUAUUGGGCGGAUGUCGCGCCGACAGAAGGCUGGUCGUGUAUAAC_GCACCAUUAAUG__ACCCAAAAAUCACGUUUGAUCCC______ .....(((((((((..((((..................)))))))))))))....................................................................... (-13.62 = -12.99 + -0.63)
| Location | 11,781,891 – 11,781,995 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 50.59 |
| Shannon entropy | 0.98166 |
| G+C content | 0.50312 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.13 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11781891 104 - 22422827 ------GGGAUAAACCGUGAUAUCUGGGU--CAUUCAGGGUGCUGCUAUACACGACCAGCCUUCUGUCGGCGCGACAUCCUCCCAAGAG--------UCUUGGGAACCGGAUGUCGCUAC-- ------((......))(((((((((((..--......(((((.(((......((((.((....))))))..))).)))))(((((((..--------.))))))).)))))))))))...-- ( -41.00, z-score = -2.65, R) >droSim1.chrX 9082224 104 - 17042790 ------GGGAUAAACCGUGAUAUCUGGGC--CAUUUAGGGUGCUGCUAGACAUGACCAGCCUUCUGUCGGCGCGACAUCCUCCCAAGAG--------UCUUGGGAACCGGAUGUCGCUAC-- ------((......))(((((((((((..--......(((((.(((..((((.((.......))))))...))).)))))(((((((..--------.))))))).)))))))))))...-- ( -42.40, z-score = -2.88, R) >droSec1.super_2776 378 104 - 2999 ------GGGAUAAACCGUGAUAUCUGGGC--CAUUUAGGGUGCUGCUAGACAUGACCAGCCUUCUGUCGGCGCGACAUCCUCCCAAGAG--------UCUUGGGAACCGGAUGUCGCUAC-- ------((......))(((((((((((..--......(((((.(((..((((.((.......))))))...))).)))))(((((((..--------.))))))).)))))))))))...-- ( -42.40, z-score = -2.88, R) >droYak2.chrX 6854945 112 + 21770863 ------GGGAUAAACCGUGAUUUCUGGGC--CAUUUGGGAUGCUGCUUGACGCGGCCAGCCUUCUGUCGGCGCGACAUCCUCCCAAGAGUGCUGGAAUCUUGGGUACCGGAUGUCGCUAC-- ------((......))...........((--(((..(((..(((((.....)))))...)))...)).)))(((((((((.(((((((.........)))))))....)))))))))...-- ( -45.10, z-score = -2.07, R) >droEre2.scaffold_4690 15331120 112 + 18748788 ------GGGAUCAGCCGUGAUUUCUAGGC--CAUUGGAAAUGCUGCUGAACGCGGCCAGCCUUCUGUCGGCGCGACAUCCUCCCAAGAGUGCAAGAGUCUUGGGAAACGGAUGUCGCUAC-- ------(.(.(((((.((.((((((((..--..)))))))))).))))).).).(((.((.....)).)))(((((((((((((((((.(.....).))))))))...)))))))))...-- ( -50.00, z-score = -3.64, R) >droAna3.scaffold_12948 293408 101 + 692136 ------GAAAUGGAACGCGAUGUCCGGGU--CAAUAGUGAUGCAGUUUAACAAGAUCGUCUUUGGAUUGGCGCGACAUCCGCCCAAUAA----------GUGGG-ACUGGACAUCGCUAC-- ------..........((((((((((.((--((....)))).)..............(((((...(((((.(((.....))))))))..----------..)))-)).)))))))))...-- ( -36.50, z-score = -2.85, R) >dp4.chrXL_group1e 8000602 105 + 12523060 AAGAUAUGAAUCUGAUCUGGGAGUCAUGU--UAUGGGUGCUGC--UUAAUCUGACUCAUCCUGGGAUAUGCGCGACAUCCGUUCAUUAG-----------UUGGAACUGGAUGUCGCUAC-- .((((....))))..((..(((((((.((--((.((......)--))))).))))...)))..))......(((((((((((((.....-----------...)))).)))))))))...-- ( -33.50, z-score = -2.02, R) >droPer1.super_17 467546 92 + 1930428 --------------AUGUGGGAGUCAUGU--CAUGAAUGCUGC-AUUAUUCUGACUCAUCCUGAAAUAUGCGCGACAUCCGUACAUUAG-----------UGAAAACUAGAUGUCGCUAC-- --------------....((((.....((--((.(((((....-..)))))))))...)))).........((((((((.......(((-----------(....))))))))))))...-- ( -26.51, z-score = -2.03, R) >droWil1.scaffold_181096 10677489 107 + 12416693 ------GAGAUGAAUGACAAUUUUAGAGUUUUUUUAGGGGAAAAUGUCGGCAACACUAGUUUGUGA-ACACCUGACAUCCGCGGAGAGA--------UUAACUUAACUGUAUUUAGUUCUUC ------(((.((((((..........(((((((((.(.(((...((((((...(((......))).-....))))))))).))))))))--------))..........)))))).)))... ( -21.55, z-score = -0.21, R) >droVir3.scaffold_12472 135760 95 - 763072 ------GAGAUCUAAUACCGAUAUGGAGU---AUUUAUGAA----UGCCAUUCGACUACGCUGUAGUUCA-GCGACGACCGAGCGUU----------UUUG-ACUCAUGGACGCCGCUGU-- ------............(((.((((.((---........)----).)))))))(((((...))))).((-(((.((.(((((((..----------..))-.)))..)).)).))))).-- ( -20.50, z-score = 0.46, R) >droMoj3.scaffold_6308 1937850 95 + 3356042 ------GAGAUCUAACACAGAUAUGGACC--AUUCACUAUU----UGCUUUAAGACUACGCUG-GGGUCA-GCGGCGACUGAGCA-U----------UUGGUACUCUAAGUCGCCGCUAC-- ------..(((((....((((((((((..--.)))).))))----)).....((......)).-)))))(-((((((((((((..-.----------......)))..))))))))))..-- ( -27.20, z-score = -1.89, R) >droGri2.scaffold_15203 9922994 97 + 11997470 ------GAGAUCCUAUGCAAAUAUGGAGC--GGUUAAUUCU----UGCUUUACGCCAACGCUGUAGUUCA-GCGGCGAACGAGCAAA----------UUUAUGCUCAAGGACGCCGCUGC-- ------....((((((....))).)))((--(((...((((----(......((((...((((.....))-))))))...(((((..----------....)))))))))).)))))...-- ( -31.30, z-score = -1.55, R) >consensus ______GGGAUCAAACGUGAUUUCUGGGC__CAUUAAUGAUGC_GUUAGACACGACCAGCCUGCUGUCGGCGCGACAUCCGCCCAAGAG________UCUUGGGAACCGGAUGUCGCUAC__ .......................................................................(((((((((............................)))))))))..... (-11.14 = -11.13 + -0.01)

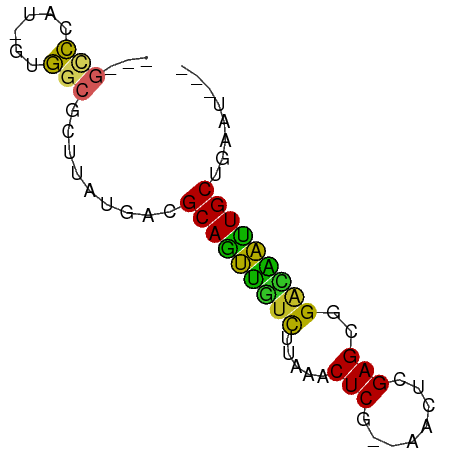
| Location | 11,781,995 – 11,782,057 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.59804 |
| G+C content | 0.47625 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -16.40 |
| Energy contribution | -14.53 |
| Covariance contribution | -1.87 |
| Combinations/Pair | 1.73 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
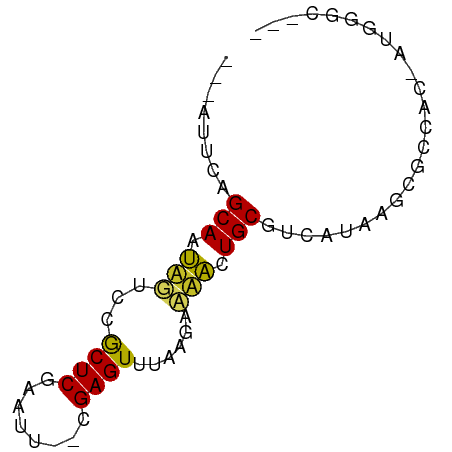
>dm3.chrX 11781995 62 + 22422827 ---GCCCAU-GUGGCGCUUAUGACGCAGUUGUCUUAAACUCG--AACUCGAGCGGGCAAUUGCUGAUU--- ---(((...-..))).........((((((((((....((((--....)))).)))))))))).....--- ( -21.10, z-score = -2.10, R) >droSim1.chrX 9082328 62 + 17042790 ---GCCCAC-GUGGCACUUAUGACGCAGUUGUCUUAAACUCG--GACUCGAGCGGGCAAUUGCUGAAU--- ---(((...-..))).........((((((((((....((((--....)))).)))))))))).....--- ( -20.20, z-score = -1.88, R) >droSec1.super_2776 482 62 + 2999 ---GCCCAC-GUGGCACUUAUGACGCAGUUGUCUUAAACUCG--GACUCGAGCGGGCAAUUGCUGAAU--- ---(((...-..))).........((((((((((....((((--....)))).)))))))))).....--- ( -20.20, z-score = -1.88, R) >droYak2.chrX 6855057 62 - 21770863 ---GCCCAU-GUGGCGCAUAUGACGCAGUUGCCUUAAACUCG--GAUUCGAGCGGACAACUGCUGAAU--- ---((((..-..)).)).......(((((((((.....((((--....)))).)).))))))).....--- ( -19.80, z-score = -1.77, R) >droEre2.scaffold_4690 15331232 62 - 18748788 ---GCCCAU-GUGGCGCAUAUGACGCAGUUGUUUUAAACUCG--UACUCGAGCGGACAACUGCUGAAU--- ---((((..-..)).)).......((((((((((....((((--....)))).)))))))))).....--- ( -21.20, z-score = -2.61, R) >droAna3.scaffold_12948 293509 60 - 692136 ---GUCCCU-GUGGCGUUUAUGACGCAGUGUUCCUAAACUCU--AACCUGAGCGGAGUACUGCCGA----- ---(((...-..))).........((((((((((....(((.--.....))).))))))))))...----- ( -15.60, z-score = -0.98, R) >dp4.chrXL_group1e 8000707 62 - 12523060 ---GCCCAUCGUGGCACUAAUGACGCAGCAUUCUAAAACUCA--AAUUUGAGCGGAUUGCUGCGGAU---- ---(((......)))........(((((((.(((....((((--....)))).))).)))))))...---- ( -21.70, z-score = -3.05, R) >droVir3.scaffold_12472 135855 65 + 763072 GCAAGAU-UUUUUGCGCAUAUGACGCAGCAUUCAUAAACUCAACGCU--GAGUGGAAUGUUGCUGAAU--- (((((..-..))))).........(((((((((....(((((....)--)))).))))))))).....--- ( -21.90, z-score = -3.00, R) >droMoj3.scaffold_6308 1937945 68 - 3356042 --GCAAUUCAUUUGCGCUUAUGACGCAAUAUUCAUAAACUCAUCAUUU-GAGUGGAAUAUUGCUGAUACAU --((((.....)))).........(((((((((....(((((.....)-)))).)))))))))........ ( -20.80, z-score = -4.05, R) >droGri2.scaffold_15203 9923091 66 - 11997470 GCAGCAUAUUUUUGCGCAUAUGACGCAAUAUUCAUAAACUCUUUAAUUCGAGUGGGAUAUUGCCGA----- ((.(((......))))).......(((((((((....((((........)))).)))))))))...----- ( -19.40, z-score = -3.05, R) >consensus ___GCCCAU_GUGGCGCUUAUGACGCAGUUGUCUUAAACUCG__AACUCGAGCGGACAAUUGCUGAAU___ ...(((......))).........(((((((((.....(((........)))..)))))))))........ (-16.40 = -14.53 + -1.87)
| Location | 11,781,995 – 11,782,057 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.59804 |
| G+C content | 0.47625 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -7.10 |
| Energy contribution | -6.17 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
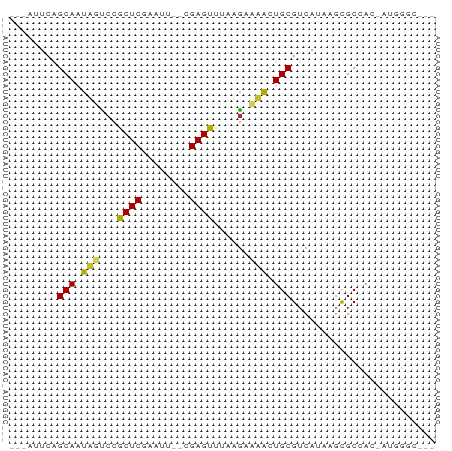
>dm3.chrX 11781995 62 - 22422827 ---AAUCAGCAAUUGCCCGCUCGAGUU--CGAGUUUAAGACAACUGCGUCAUAAGCGCCAC-AUGGGC--- ---...........(((((((((....--)))))...........((((.....))))...-..))))--- ( -16.60, z-score = -1.08, R) >droSim1.chrX 9082328 62 - 17042790 ---AUUCAGCAAUUGCCCGCUCGAGUC--CGAGUUUAAGACAACUGCGUCAUAAGUGCCAC-GUGGGC--- ---...........(((((((((....--)))))....(((......)))...........-..))))--- ( -14.90, z-score = -0.20, R) >droSec1.super_2776 482 62 - 2999 ---AUUCAGCAAUUGCCCGCUCGAGUC--CGAGUUUAAGACAACUGCGUCAUAAGUGCCAC-GUGGGC--- ---...........(((((((((....--)))))....(((......)))...........-..))))--- ( -14.90, z-score = -0.20, R) >droYak2.chrX 6855057 62 + 21770863 ---AUUCAGCAGUUGUCCGCUCGAAUC--CGAGUUUAAGGCAACUGCGUCAUAUGCGCCAC-AUGGGC--- ---.....(((((((((.(((((....--)))))....))))))))).........(((..-...)))--- ( -22.80, z-score = -2.83, R) >droEre2.scaffold_4690 15331232 62 + 18748788 ---AUUCAGCAGUUGUCCGCUCGAGUA--CGAGUUUAAAACAACUGCGUCAUAUGCGCCAC-AUGGGC--- ---.....((((((((..(((((....--))))).....)))))))).........(((..-...)))--- ( -19.50, z-score = -2.14, R) >droAna3.scaffold_12948 293509 60 + 692136 -----UCGGCAGUACUCCGCUCAGGUU--AGAGUUUAGGAACACUGCGUCAUAAACGCCAC-AGGGAC--- -----...(((((..(((((((.....--.))))...)))..)))))..........((..-..))..--- ( -14.40, z-score = -0.67, R) >dp4.chrXL_group1e 8000707 62 + 12523060 ----AUCCGCAGCAAUCCGCUCAAAUU--UGAGUUUUAGAAUGCUGCGUCAUUAGUGCCACGAUGGGC--- ----...(((((((.((.(((((....--)))))....)).)))))))........(((......)))--- ( -19.60, z-score = -2.07, R) >droVir3.scaffold_12472 135855 65 - 763072 ---AUUCAGCAACAUUCCACUC--AGCGUUGAGUUUAUGAAUGCUGCGUCAUAUGCGCAAAAA-AUCUUGC ---.....(((((((((.((((--......))))....))))).(((((.....)))))....-...)))) ( -13.80, z-score = -0.47, R) >droMoj3.scaffold_6308 1937945 68 + 3356042 AUGUAUCAGCAAUAUUCCACUC-AAAUGAUGAGUUUAUGAAUAUUGCGUCAUAAGCGCAAAUGAAUUGC-- ........(((((((((.((((-(.....)))))....))))))))).........((((.....))))-- ( -16.00, z-score = -1.72, R) >droGri2.scaffold_15203 9923091 66 + 11997470 -----UCGGCAAUAUCCCACUCGAAUUAAAGAGUUUAUGAAUAUUGCGUCAUAUGCGCAAAAAUAUGCUGC -----.(((((.......((((........)))).........((((((.....)))))).....))))). ( -15.00, z-score = -1.74, R) >consensus ___AUUCAGCAAUAGUCCGCUCGAAUU__CGAGUUUAAGAAAACUGCGUCAUAAGCGCCAC_AUGGGC___ ........(((.(((...((((........))))......))).)))........................ ( -7.10 = -6.17 + -0.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:38 2011