| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,771,222 – 11,771,358 |
| Length | 136 |
| Max. P | 0.999404 |

| Location | 11,771,222 – 11,771,358 |
|---|---|
| Length | 136 |
| Sequences | 4 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Shannon entropy | 0.32032 |
| G+C content | 0.37400 |
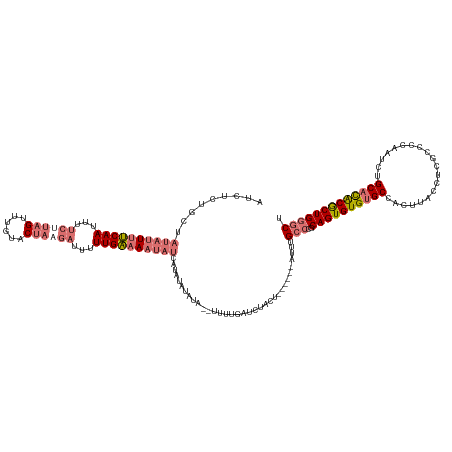
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -21.95 |
| Energy contribution | -24.33 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
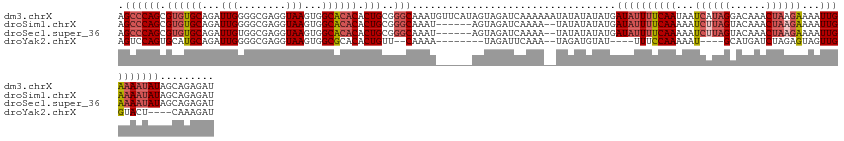
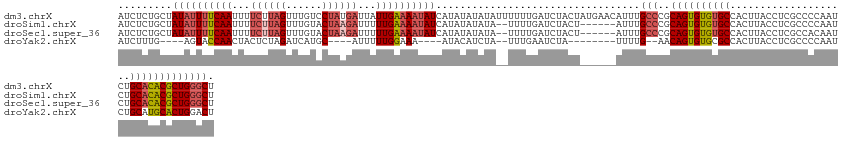
>dm3.chrX 11771222 136 + 22422827 AGCCCAGCGUGUGCAGAUUGGGGCGAGGUAAGUGGCACACACUGCGGGCAAAUGUUCAUAGUAGAUCAAAAAAUAUAUAUAUGAUAUUUUCAAUAAUCAUAGGACAAACUAAGAAAAUUGAAAAUAUAGCAGAGAU .((((((.((((((...(((........)))...)))))).))..))))...((((((((...................))))(((((((((((..((.(((......))).))..)))))))))))))))..... ( -32.01, z-score = -2.38, R) >droSim1.chrX 9071526 128 + 17042790 AGCCCAGCGUGUGCAGAUUGGGGCGAGGUAAGUGGCACACACUGCGGGCAAAU------AGUAGAUCAAAA--UAUAUAUAUGAUAUUUUCAAAAAUCUUAGUACAAACUAAGAAAAUUGAAAAUAUAGCAGAGAU .((((((.((((((...(((........)))...)))))).))..))))....------............--..........((((((((((...(((((((....)))))))...))))))))))......... ( -33.90, z-score = -3.52, R) >droSec1.super_36 444253 128 + 449511 AGCCCAGCGUGUGCAGAUUGUGGCGAGGUAAGUGGCACACACUGCGGGCAAAU------AGUAGAUCAAAA--UAUAUAUAUGAUAUUUUCAAAAAUCUUAGUACAAACUAAGAAAAUUGAAAAUAUAGCAGAGAU .((((.(((((((.....(((.((.......)).)))))))).))))))....------............--..........((((((((((...(((((((....)))))))...))))))))))......... ( -35.10, z-score = -3.81, R) >droYak2.chrX 6844080 112 - 21770863 AGUCCAGUGCAUGCAGAUUGGGGCGAGGUAAGUGGCGCACACUGUU--CAAAA--------UAGAUUCAAA--UAGAUGUAU----UUUCCAAAAAU----GCAUGAUCUAGAGUAGUUGGUACU----CAAAGAU .....((..((((((..((((...(((...((((.....)))).))--)((((--------((.(((....--..))).)))----)))))))...)----)))))..)).(((((.....))))----)...... ( -25.30, z-score = -0.41, R) >consensus AGCCCAGCGUGUGCAGAUUGGGGCGAGGUAAGUGGCACACACUGCGGGCAAAU______AGUAGAUCAAAA__UAUAUAUAUGAUAUUUUCAAAAAUCUUAGUACAAACUAAGAAAAUUGAAAAUAUAGCAGAGAU .((((((.((((((...(((........)))...)))))).)))..)))..................................((((((((((...((((((......))))))...))))))))))......... (-21.95 = -24.33 + 2.37)
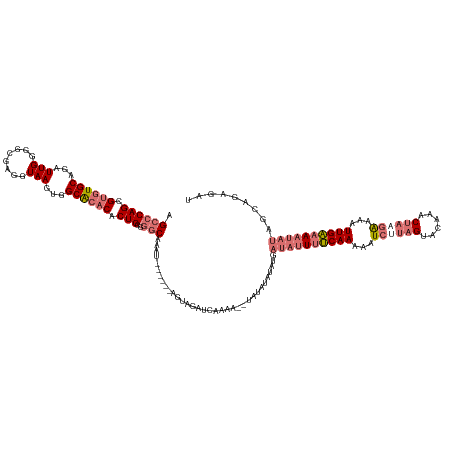
| Location | 11,771,222 – 11,771,358 |
|---|---|
| Length | 136 |
| Sequences | 4 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Shannon entropy | 0.32032 |
| G+C content | 0.37400 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -22.66 |
| Energy contribution | -24.85 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.08 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11771222 136 - 22422827 AUCUCUGCUAUAUUUUCAAUUUUCUUAGUUUGUCCUAUGAUUAUUGAAAAUAUCAUAUAUAUAUUUUUUGAUCUACUAUGAACAUUUGCCCGCAGUGUGUGCCACUUACCUCGCCCCAAUCUGCACACGCUGGGCU ..........((((((((((..((.(((......))).))..))))))))))(((((.....(((....)))....)))))......((((..(((((((((....................))))))))))))). ( -31.75, z-score = -3.91, R) >droSim1.chrX 9071526 128 - 17042790 AUCUCUGCUAUAUUUUCAAUUUUCUUAGUUUGUACUAAGAUUUUUGAAAAUAUCAUAUAUAUA--UUUUGAUCUACU------AUUUGCCCGCAGUGUGUGCCACUUACCUCGCCCCAAUCUGCACACGCUGGGCU .........((((((((((...(((((((....)))))))...))))))))))..........--............------....((((..(((((((((....................))))))))))))). ( -35.05, z-score = -5.59, R) >droSec1.super_36 444253 128 - 449511 AUCUCUGCUAUAUUUUCAAUUUUCUUAGUUUGUACUAAGAUUUUUGAAAAUAUCAUAUAUAUA--UUUUGAUCUACU------AUUUGCCCGCAGUGUGUGCCACUUACCUCGCCACAAUCUGCACACGCUGGGCU .........((((((((((...(((((((....)))))))...))))))))))..........--............------....((((..(((((((((....................))))))))))))). ( -35.05, z-score = -5.37, R) >droYak2.chrX 6844080 112 + 21770863 AUCUUUG----AGUACCAACUACUCUAGAUCAUGC----AUUUUUGGAAA----AUACAUCUA--UUUGAAUCUA--------UUUUG--AACAGUGUGCGCCACUUACCUCGCCCCAAUCUGCAUGCACUGGACU ((((..(----((((.....))))).))))(((((----(...((((...----..(((.((.--((..((....--------.))..--)).)))))(((..........))).))))..))))))......... ( -21.50, z-score = -1.43, R) >consensus AUCUCUGCUAUAUUUUCAAUUUUCUUAGUUUGUACUAAGAUUUUUGAAAAUAUCAUAUAUAUA__UUUUGAUCUACU______AUUUGCCCGCAGUGUGUGCCACUUACCUCGCCCCAAUCUGCACACGCUGGGCU .........((((((((((...((((((......))))))...))))))))))..................................(((..((((((((((....................))))))))))))). (-22.66 = -24.85 + 2.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:33 2011