
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,716,989 – 11,717,117 |
| Length | 128 |
| Max. P | 0.630212 |

| Location | 11,716,989 – 11,717,088 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 60.51 |
| Shannon entropy | 0.75607 |
| G+C content | 0.30770 |
| Mean single sequence MFE | -9.97 |
| Consensus MFE | -4.59 |
| Energy contribution | -4.62 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.49 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
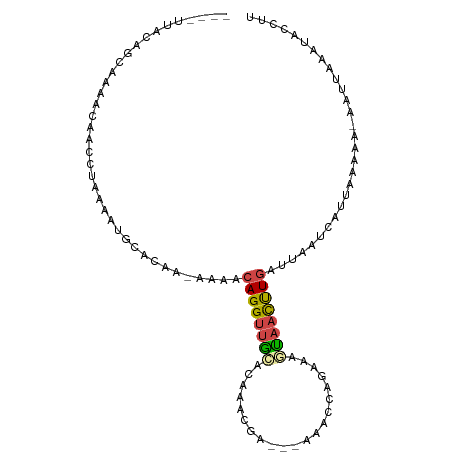
>dm3.chrX 11716989 99 - 22422827 UUAAAAAAAGCAAAACAACCUAAAAUGCACAAAAAAACAGGUUGCACAAGCGA---UAAC-CAGAAGUAACUUGAUUAAUCAUUUAAAAAAAUUAAAUACCUU (((((....(((.............))).........((((((((........---....-.....))))))))........)))))................ ( -8.65, z-score = -0.62, R) >droSim1.chrU 8530902 91 + 15797150 ----UUACAGCAAAACAACCUAAAAUGCACAA-AAAACAGGUUGCACAAACAAGCGAUACCAGAAAGUAACUUGAUUAAUCAUUAAA-------AAAUACCUU ----.....(((.............)))....-....((((((((.....................)))))))).............-------......... ( -8.12, z-score = -0.53, R) >droSec1.super_36 390481 97 - 449511 ----GUACAGCAAAACAACCUAAAAUGCACAA-AAAACAGGUUGCACAAACAAGCGAUACCAGAAAGUAACUUGAUUAAUCAUUAAAAA-AAUUAAAUACCUU ----.....(((.............)))....-....((((((((.....................))))))))...............-............. ( -8.12, z-score = -0.10, R) >droYak2.chrX 6788529 77 + 21770863 ----UUACAGCAAAACAACCUGAAAUGCACCG-AAAACAGGUUGC---------------------UCAACUUGAUGUAUCAUUAAAAAUAAUUAAACACUUU ----.((((.(((..(((((((..........-....))))))).---------------------.....))).))))........................ ( -9.74, z-score = -1.20, R) >droEre2.scaffold_4690 15269104 95 + 18748788 ----UUACAGCAAAACAAUCUAAAAUGUAGCGAAAAACAGGUUGCUAAUGCGU---UAACAUGAAAUCAACUUGAUUUAUCAUUUAUAA-AAUUAAAUACCUU ----.....((...(((........))).))((.((((((((((...(((...---...))).....))))))).))).))(((((...-...)))))..... ( -10.20, z-score = 0.05, R) >droAna3.scaffold_489 1617 97 + 2139 ----AAAACGCAGGAGUGCCCGAUAUGACCCACUACCAAGGCUAUAUCUUUGCCAAAAAUCAUCCGAUUCUCUAGCGGAAUACCUUAAAAGAUUAAAAAUC-- ----....(((.((((.((..((((((.((.........)).))))))...))....((((....)))))))).)))........................-- ( -15.00, z-score = -0.56, R) >consensus ____UUACAGCAAAACAACCUAAAAUGCACAA_AAAACAGGUUGCACAAACGA___AAACCAGAAAGUAACUUGAUUAAUCAUUAAAAA_AAUUAAAUACCUU .....................................((((((((.....................))))))))............................. ( -4.59 = -4.62 + 0.03)

| Location | 11,717,015 – 11,717,117 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 60.57 |
| Shannon entropy | 0.73099 |
| G+C content | 0.37315 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -8.41 |
| Energy contribution | -9.03 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11717015 102 + 22422827 AAUCAAGUU-ACUUCUGGUUAUCGCUUG----UGCAACCUGUUUUUUUGUGCAUUUUAGGUUGUUUUGCUUUUUUUAAUUGUACCUAGCGAUGAGUCAAUCCACUUA- ....((((.-.....(((((((((((.(----(((((..(((........)))....((((......)))).......))))))..)))))))).)))....)))).- ( -18.40, z-score = -0.82, R) >droSim1.chrU 8530921 101 - 15797150 AAUCAAGUU-ACUUUCUGGUAUCGCUUGUUUGUGCAACCUGUUUUUU-GUGCAUUUUAGGUUGUUUUGCUGU----AAUUGUACUUAGCGAUGAGUCAACCCACUUA- ...((((((-(((....))))..)))))..((..(((........))-)..)).....(((((..((((((.----.........)))))).....)))))......- ( -20.50, z-score = -0.71, R) >droSec1.super_36 390506 101 + 449511 AAUCAAGUU-ACUUUCUGGUAUCGCUUGUUUGUGCAACCUGUUUUUU-GUGCAUUUUAGGUUGUUUUGCUGU----ACUUGUACUUAGCGAUGAAUCAACCCACUUA- ...((((((-(((....))))..)))))..((..(((........))-)..)).....(((((..(((((((----(....)))..))))).....)))))......- ( -21.00, z-score = -1.13, R) >droYak2.chrX 6788555 81 - 21770863 -------------------CAUCAAGUU---GAGCAACCUGUUUUCG-GUGCAUUUCAGGUUGUUUUGCUGU----AAUUGUACUUUGCAAUGAGUCAAUUCGCAUAU -------------------.....(((.---((((((((((......-........)))))))))).)))((----(((((.((((......))))))))).)).... ( -21.14, z-score = -1.84, R) >droEre2.scaffold_4690 15269129 99 - 18748788 AAUCAAGUUGAUUUCAUGUUAACGCAUU----AGCAACCUGUUUUUCGCUACAUUUUAGAUUGUUUUGCUGU----AAUUGUACUUUGCGAUGAGUCAAUCCACAUA- ......((((((.....))))))(((..----(((((.(((...............))).))))).)))(((----.((((.((((......))))))))..)))..- ( -15.46, z-score = 0.35, R) >droAna3.scaffold_489 1647 85 - 2139 ---AGAAUCGGAUGAUUUUUGGCAAAGA----UAUAGCCUUGGUAGUGGGUCAUAUCGGGCACUCCUGCGUU----UUCCAAGAUUUUCAAGGGGA------------ ---((((((....))))))..((...((----(((..(((.......)))..)))))..)).(((((.....----.((...))......))))).------------ ( -18.60, z-score = 0.80, R) >consensus AAUCAAGUU_ACUUCCUGUUAUCGCUUG____UGCAACCUGUUUUUU_GUGCAUUUUAGGUUGUUUUGCUGU____AAUUGUACUUAGCGAUGAGUCAAUCCACUUA_ ...................(((((((.......((((((((...............))))))))..(((...........)))...)))))))............... ( -8.41 = -9.03 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:29 2011