
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,711,721 – 11,711,835 |
| Length | 114 |
| Max. P | 0.500000 |

| Location | 11,711,721 – 11,711,835 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 62.05 |
| Shannon entropy | 0.74500 |
| G+C content | 0.44933 |
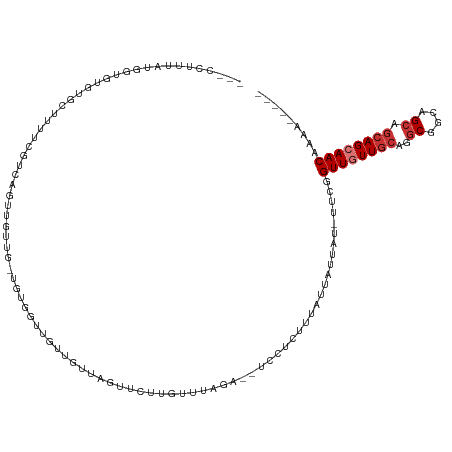
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
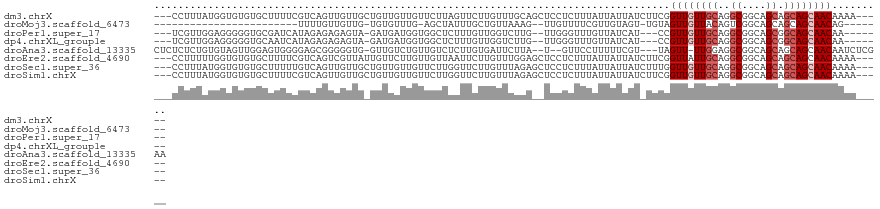
>dm3.chrX 11711721 114 - 22422827 ---CCUUUAUGGUGUGUGCUUUUCGUCAGUUGUUGCUGUUGUUGUUCUUAGUUCUUGUUUGCAGCUCCUCUUUAUUAUUAUCUUCGGUUGUUGCAGGCGGCAGCAGCAGCAACAAAA----- ---((.....)).................((((((((((((((((..........((((((((((.((.................))..))))))))))))))))))))))))))..----- ( -35.73, z-score = -3.37, R) >droMoj3.scaffold_6473 638303 86 - 16943266 ------------------------UUUUGUUGUUG-UGUGUUUG-AGCUAUUUGCUGUUAAAG--UUGUUUUCGUUGUAGU-UGUAGUUGUUACAGUCGGCAGCAGCAGCAACAG------- ------------------------...((((((((-(.......-(((.....))).......--........(((((.((-(((((...)))))).).))))).))))))))).------- ( -21.80, z-score = -1.16, R) >droPer1.super_17 1081081 106 + 1930428 ---UCGUUGGAGGGGGUGCGAUCAUAGAGAGAGUA-GAUGAUGGUGGCUCUUUGUUGGUCUUG--UUGGGUUUGUUAUCAU---CCGUUGUUGCAGGCGGCAGCGGCAGCAACAA------- ---......((((((.(((.(((((..........-.))))).))).)))))).......(((--((((((.....)))..---(((((((((....)))))))))))))))...------- ( -30.70, z-score = -0.32, R) >dp4.chrXL_group1e 8609554 106 + 12523060 ---UCGUUGGAGGGGGUGCAAUCAUAGAGAGAGUA-GAUGAUGGUGGCUCUUUGUUGGUCUUG--UUGGGUUUGUUAUCAU---CCGUUGUUGCAGGCGGCAGCGGCAGCAACAA------- ---........(..(.(((................-((((((((..((((.............--..))).)..)))))))---)((((((((....))))))))))).)..)..------- ( -28.96, z-score = 0.09, R) >droAna3.scaffold_13335 763374 113 - 3335858 CUCUCUCUGUGUAGUUGGAGUGGGGAGCGGGGGUG-GUUGUCUGUUGUCUCUUGUGAUUCUUA--U--GUUCCUUUUUCGU---UAGUU-UUGGAGGCGGCAGCAGCAGCAACAAUCUCGAA (((.(((((......))))).)))...(((((...-(((((((((((((.(((.(((..((.(--(--(.........)))---.))..-))).))).))))))))..)))))..))))).. ( -35.30, z-score = -1.34, R) >droEre2.scaffold_4690 15262038 114 + 18748788 ---CCUUUUUGGUGUGUGCUUUUCGUCAGUCGUUAUUGUUCUUGUUGUUAAUUCUUGUUUGGAGCUCCUCUUUAUUAUUAUCUUCGGUUAUUGCAGGCGGCAGCAGCAGCAACAAAA----- ---.....(((..(.........)..)))......(((((.((((((((...(((.....)))(((((((..((.......(....).))..).))).))))))))))).)))))..----- ( -17.50, z-score = 1.87, R) >droSec1.super_36 385351 114 - 449511 ---CCUUUAUGGUGUGUGCUUUUUGUCAGUUGUUGCUGUUGUUGUUCUUGGUUCUUGUUUAGAGCUCCUCUUUAUUAUUAUCUUUGGUUGUUGCAGGCGGCAGCAGCAGCAACAAAA----- ---((.....)).................((((((((((((((((((((((((((.....))))).((.................))......)))).)))))))))))))))))..----- ( -32.33, z-score = -1.91, R) >droSim1.chrX 9023587 114 - 17042790 ---CCUUUAUGGUGUGUGCUUUUCGUCAGUUGUUGCUGUUGUUGUUCUUGGUUCUUGUUUAGAGCUCCUCUUUAUUAUUAUCUUCGGUUGUUGCAGGCGGCAGCAGCAGCAACAAAA----- ---((.....)).................((((((((((((((((((((((((((.....)))))................(....)......)))).)))))))))))))))))..----- ( -33.30, z-score = -2.41, R) >consensus ___CCUUUAUGGUGUGUGCUUUUCGUCAGUUGUUG_UGUGGUUGUUGUUAGUUCUUGUUUAGA__UCCUCUUUAUUAUUAU_UUCGGUUGUUGCAGGCGGCAGCAGCAGCAACAAAA_____ ......................................................................................((((((((..((....)).))))))))......... ( -9.94 = -10.56 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:27 2011