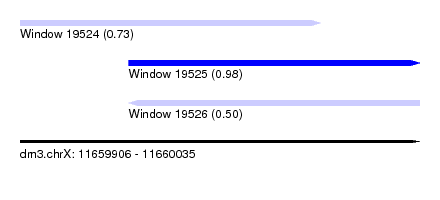
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,659,906 – 11,660,035 |
| Length | 129 |
| Max. P | 0.979064 |

| Location | 11,659,906 – 11,660,003 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 63.80 |
| Shannon entropy | 0.72506 |
| G+C content | 0.52125 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -10.44 |
| Energy contribution | -10.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732628 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
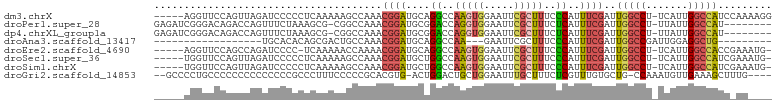
>dm3.chrX 11659906 97 + 22422827 -----AGGUUCCAGUUAGAUCCCCCUCAAAAAGCCAAACGGAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCAUCCAAAAGG -----.((.((......)).)).(((......((((.....(((.(((((((.((((((..........)))))).)))))))-.)))))))........))) ( -21.54, z-score = 0.64, R) >droPer1.super_28 412208 93 - 1111753 GAGAUCGGGACAGACCAGUUUCUAAAGCG-CGGCCAAACGGAUGCGGACCAGGUGGAAUUCGCUUUCUCAUUUCGAUUGGCCU-UUAUUGGCCAU-------- (((((.((((.((.(.((((((.....((-((.((....)).))))........)))))).))).)))))))))...(((((.-.....))))).-------- ( -26.72, z-score = -0.23, R) >dp4.chrXL_group1a 6887832 93 - 9151740 GAGAUCGGGACAGACCAGUUUCUAAAGCG-CGGCCAAACGGAUGCGGACCAGGUGGAAUUCGCUUUCUCAUUUCGAUUGGCCU-UUAUUGGCCAU-------- (((((.((((.((.(.((((((.....((-((.((....)).))))........)))))).))).)))))))))...(((((.-.....))))).-------- ( -26.72, z-score = -0.23, R) >droAna3.scaffold_13417 1407997 73 + 6960332 ------------------UGCACACAGCGACUGCCAAACGGAUGCAGGCCAA---GAAUUCGCUUUCCCAUUUCGAUUGGCCGAUUGGAGGCUG--------- ------------------.((...(((((.(((.....))).))).(((((.---(((.............)))...)))))...))...))..--------- ( -16.72, z-score = 0.79, R) >droEre2.scaffold_4690 15208781 95 - 18748788 -----AGGUUCCAGCCAGAUCCCC-UCAAAAACCAAAACGGAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCACCGAAAUG- -----.((((((((((.(((((..-..............)))).).)))....)))))))........(((((((..(((((.-.....))))).)))))))- ( -27.09, z-score = -1.53, R) >droSec1.super_36 335510 96 + 449511 -----UGGUUCCAGUUAGAUCCCCCUCAAAAAGCCAAACGGAUGCUGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCAUCGAAAUG- -----(((..(((((...((((.................))))))))))))...((((......))))(((((((((.((((.-.....)))))))))))))- ( -30.93, z-score = -2.51, R) >droSim1.chrX 8976534 96 + 17042790 -----UGGUUCCAGUUAGAUCCCCCUCAAAAAGCCAAACGGAUGCUGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCAUCGAAAUG- -----(((..(((((...((((.................))))))))))))...((((......))))(((((((((.((((.-.....)))))))))))))- ( -30.93, z-score = -2.51, R) >droGri2.scaffold_14853 8345402 95 - 10151454 --GCCCCUGCCCCCCCCCCCCCCGCCCUUUCCCCCGCACGUG-ACUGGACUGCUGGAAUUUGCUUUCUCGUUUGUGCUG-CCAAAUGUUGAAAGCUUUG---- --..............................((.(((.((.-.....))))).)).....((((((.((((((.....-.))))))..))))))....---- ( -17.60, z-score = -1.83, R) >consensus _____CGGUUCCAGCCAGAUCCCCCUCAAAAAGCCAAACGGAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU_UCAUUGGCCAUC_AA____ ......................................((((....((..(((((.....)))))..))..))))..(((((.......)))))......... (-10.44 = -10.68 + 0.24)
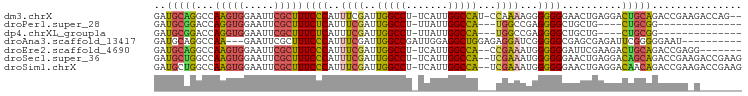
| Location | 11,659,941 – 11,660,035 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.54608 |
| G+C content | 0.55508 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
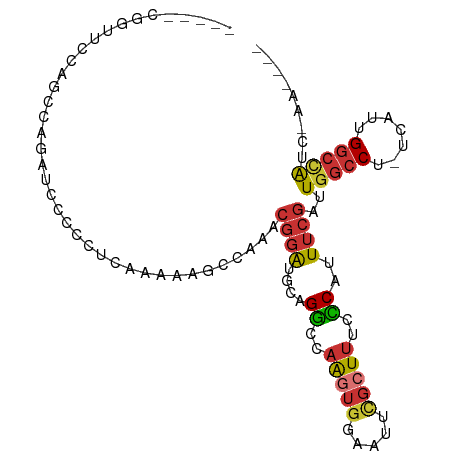
>dm3.chrX 11659941 94 + 22422827 GAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCAU-CCAAAAGGGGGGGAACUGAGGACUGCAGACCGAAGACCAG-- ..(((((.((.(((........((..(((.(((.(((.((((.-.....))))))-).))).)))..)).)))..)).))))).............-- ( -32.00, z-score = -0.94, R) >droPer1.super_28 412247 76 - 1111753 GAUGCGGACCAGGUGGAAUUCGCUUUCUCAUUUCGAUUGGCCU-UUAUUGGCCA---UGGCCGAGGGGCUGCUG----CUGCGG-------------- ....(((.((((((((((......))).)))).....(((((.-.....)))))---)))))).(.(((....)----)).)..-------------- ( -24.20, z-score = 0.24, R) >dp4.chrXL_group1a 6887871 76 - 9151740 GAUGCGGACCAGGUGGAAUUCGCUUUCUCAUUUCGAUUGGCCU-UUAUUGGCCA---UGGCCGAGGGGCUGCUG----CUGCGG-------------- ....(((.((((((((((......))).)))).....(((((.-.....)))))---)))))).(.(((....)----)).)..-------------- ( -24.20, z-score = 0.24, R) >droAna3.scaffold_13417 1408019 85 + 6960332 GAUGCAGGCCAA---GAAUUCGCUUUCCCAUUUCGAUUGGCCGAUUGGAGGCUGGAGAGGAUCGGGGGCGAGCGAGAUUCGGGGGAAU---------- ........((..---((((((((((((((..(((..(..(((.......)))..)...)))...)))).)))))).))))..))....---------- ( -29.60, z-score = -0.69, R) >droEre2.scaffold_4690 15208815 88 - 18748788 GAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCA--CCGAAAUGGGGGGAUUCGAAGACUGCAGACCGAGG------- ..(((((.((....))..((((.(..(((((((((..(((((.-.....)))))--.)))))))))..)...))))..)))))........------- ( -36.80, z-score = -3.02, R) >droSec1.super_36 335545 95 + 449511 GAUGCUGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCA--UCGAAAUGGGGGGAACUGAGGACAGCAGACCGAAGACCGAAG ..(((((.((..(((.....)))(..(((((((((((.((((.-.....)))))--))))))))))..)......)).)))))............... ( -40.10, z-score = -3.64, R) >droSim1.chrX 8976569 95 + 17042790 GAUGCUGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU-UCAUUGGCCA--UCGAAAUGGGGGGAACUGAGGACAACAGACCGAAGACCGAAG ...(((.....)))((..((((.(..(((((((((((.((((.-.....)))))--))))))))))..)..(((.......)))..))))..)).... ( -36.40, z-score = -2.92, R) >consensus GAUGCAGGCCAAGUGGAAUUCGCUUUCCCAUUUCGAUUGGCCU_UCAUUGGCCA___CGAAAGGGGGGGAACUGAGGACUGCAGACCGA_G_______ ..(((((...(((((.....)))))((((..((((..(((((.......)))))...))))...))))..........)))))............... (-20.66 = -20.26 + -0.40)
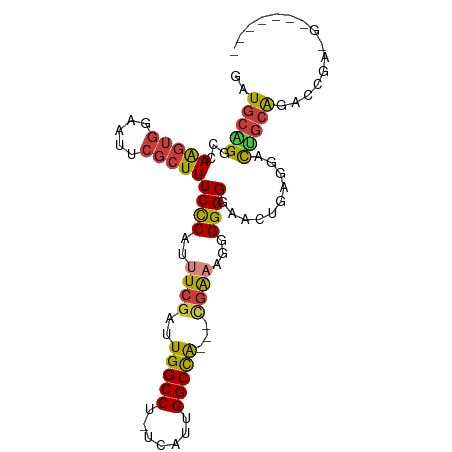
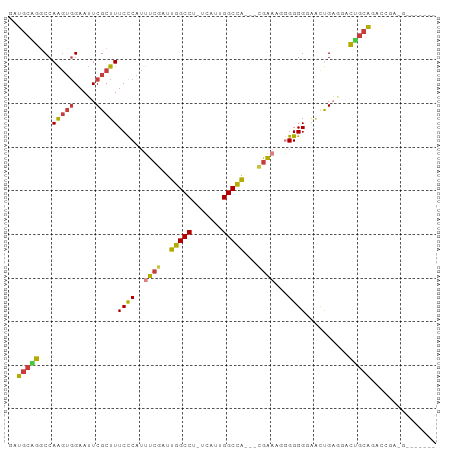
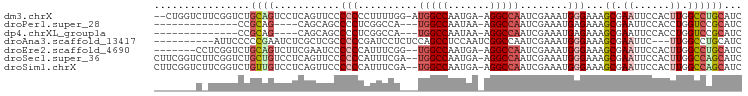
| Location | 11,659,941 – 11,660,035 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.54608 |
| G+C content | 0.55508 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -11.17 |
| Energy contribution | -11.79 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 11659941 94 - 22422827 --CUGGUCUUCGGUCUGCAGUCCUCAGUUCCCCCCCUUUUGG-AUGGCCAAUGA-AGGCCAAUCGAAAUGGGAAAGCGAAUUCCACUUGGCCUGCAUC --..((.(....).))((((.((..((((((.(((.((((((-.(((((.....-.))))).)))))).)))...).)))))......)).))))... ( -30.40, z-score = -0.91, R) >droPer1.super_28 412247 76 + 1111753 --------------CCGCAG----CAGCAGCCCCUCGGCCA---UGGCCAAUAA-AGGCCAAUCGAAAUGAGAAAGCGAAUUCCACCUGGUCCGCAUC --------------..((.(----(....)).....(((((---(((((.....-.))))).(((...........)))........))))).))... ( -18.10, z-score = 0.11, R) >dp4.chrXL_group1a 6887871 76 + 9151740 --------------CCGCAG----CAGCAGCCCCUCGGCCA---UGGCCAAUAA-AGGCCAAUCGAAAUGAGAAAGCGAAUUCCACCUGGUCCGCAUC --------------..((.(----(....)).....(((((---(((((.....-.))))).(((...........)))........))))).))... ( -18.10, z-score = 0.11, R) >droAna3.scaffold_13417 1408019 85 - 6960332 ----------AUUCCCCCGAAUCUCGCUCGCCCCCGAUCCUCUCCAGCCUCCAAUCGGCCAAUCGAAAUGGGAAAGCGAAUUC---UUGGCCUGCAUC ----------....((..((((.(((((..(((.((((........(((.......)))..))))....)))..)))))))))---..))........ ( -19.30, z-score = -0.13, R) >droEre2.scaffold_4690 15208815 88 + 18748788 -------CCUCGGUCUGCAGUCUUCGAAUCCCCCCAUUUCGG--UGGCCAAUGA-AGGCCAAUCGAAAUGGGAAAGCGAAUUCCACUUGGCCUGCAUC -------........(((((..((((......((((((((((--(((((.....-.)))).)))))))))))....))))..((....)).))))).. ( -33.40, z-score = -2.91, R) >droSec1.super_36 335545 95 - 449511 CUUCGGUCUUCGGUCUGCUGUCCUCAGUUCCCCCCAUUUCGA--UGGCCAAUGA-AGGCCAAUCGAAAUGGGAAAGCGAAUUCCACUUGGCCAGCAUC ....((.(....).))((((.((..((((((.((((((((((--(((((.....-.)))).)))))))))))...).)))))......)).))))... ( -36.80, z-score = -3.50, R) >droSim1.chrX 8976569 95 - 17042790 CUUCGGUCUUCGGUCUGUUGUCCUCAGUUCCCCCCAUUUCGA--UGGCCAAUGA-AGGCCAAUCGAAAUGGGAAAGCGAAUUCCACUUGGCCAGCAUC ....((((...((.(((.......)))..)).((((((((((--(((((.....-.)))).)))))))))))................))))...... ( -34.60, z-score = -3.01, R) >consensus _______C_UCGGUCUGCAGUCCUCAGUUCCCCCCAUUUCG___UGGCCAAUGA_AGGCCAAUCGAAAUGGGAAAGCGAAUUCCACUUGGCCUGCAUC ................((((............(((.........(((((.......)))))........)))...((.((......)).))))))... (-11.17 = -11.79 + 0.62)
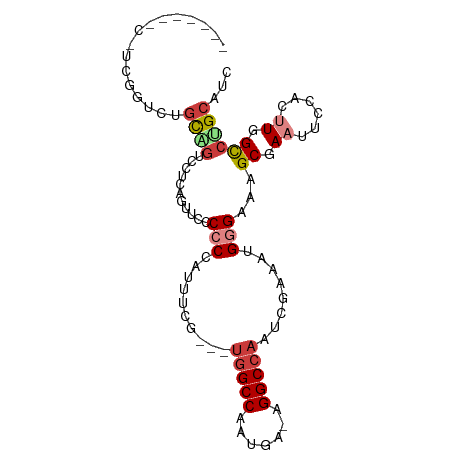
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:25 2011